Abdelmalik Taleb-Ahmed
LAMIH, University of Valenciennes, France
RAVID: Retrieval-Augmented Visual Detection: A Knowledge-Driven Approach for AI-Generated Image Identification
Aug 05, 2025Abstract:In this paper, we introduce RAVID, the first framework for AI-generated image detection that leverages visual retrieval-augmented generation (RAG). While RAG methods have shown promise in mitigating factual inaccuracies in foundation models, they have primarily focused on text, leaving visual knowledge underexplored. Meanwhile, existing detection methods, which struggle with generalization and robustness, often rely on low-level artifacts and model-specific features, limiting their adaptability. To address this, RAVID dynamically retrieves relevant images to enhance detection. Our approach utilizes a fine-tuned CLIP image encoder, RAVID CLIP, enhanced with category-related prompts to improve representation learning. We further integrate a vision-language model (VLM) to fuse retrieved images with the query, enriching the input and improving accuracy. Given a query image, RAVID generates an embedding using RAVID CLIP, retrieves the most relevant images from a database, and combines these with the query image to form an enriched input for a VLM (e.g., Qwen-VL or Openflamingo). Experiments on the UniversalFakeDetect benchmark, which covers 19 generative models, show that RAVID achieves state-of-the-art performance with an average accuracy of 93.85%. RAVID also outperforms traditional methods in terms of robustness, maintaining high accuracy even under image degradations such as Gaussian blur and JPEG compression. Specifically, RAVID achieves an average accuracy of 80.27% under degradation conditions, compared to 63.44% for the state-of-the-art model C2P-CLIP, demonstrating consistent improvements in both Gaussian blur and JPEG compression scenarios. The code will be publicly available upon acceptance.
Advancing Wheat Crop Analysis: A Survey of Deep Learning Approaches Using Hyperspectral Imaging
May 01, 2025Abstract:As one of the most widely cultivated and consumed crops, wheat is essential to global food security. However, wheat production is increasingly challenged by pests, diseases, climate change, and water scarcity, threatening yields. Traditional crop monitoring methods are labor-intensive and often ineffective for early issue detection. Hyperspectral imaging (HSI) has emerged as a non-destructive and efficient technology for remote crop health assessment. However, the high dimensionality of HSI data and limited availability of labeled samples present notable challenges. In recent years, deep learning has shown great promise in addressing these challenges due to its ability to extract and analysis complex structures. Despite advancements in applying deep learning methods to HSI data for wheat crop analysis, no comprehensive survey currently exists in this field. This review addresses this gap by summarizing benchmark datasets, tracking advancements in deep learning methods, and analyzing key applications such as variety classification, disease detection, and yield estimation. It also highlights the strengths, limitations, and future opportunities in leveraging deep learning methods for HSI-based wheat crop analysis. We have listed the current state-of-the-art papers and will continue tracking updating them in the following https://github.com/fadi-07/Awesome-Wheat-HSI-DeepLearning.
DeeCLIP: A Robust and Generalizable Transformer-Based Framework for Detecting AI-Generated Images
Apr 28, 2025Abstract:This paper introduces DeeCLIP, a novel framework for detecting AI-generated images using CLIP-ViT and fusion learning. Despite significant advancements in generative models capable of creating highly photorealistic images, existing detection methods often struggle to generalize across different models and are highly sensitive to minor perturbations. To address these challenges, DeeCLIP incorporates DeeFuser, a fusion module that combines high-level and low-level features, improving robustness against degradations such as compression and blurring. Additionally, we apply triplet loss to refine the embedding space, enhancing the model's ability to distinguish between real and synthetic content. To further enable lightweight adaptation while preserving pre-trained knowledge, we adopt parameter-efficient fine-tuning using low-rank adaptation (LoRA) within the CLIP-ViT backbone. This approach supports effective zero-shot learning without sacrificing generalization. Trained exclusively on 4-class ProGAN data, DeeCLIP achieves an average accuracy of 89.00% on 19 test subsets composed of generative adversarial network (GAN) and diffusion models. Despite having fewer trainable parameters, DeeCLIP outperforms existing methods, demonstrating superior robustness against various generative models and real-world distortions. The code is publicly available at https://github.com/Mamadou-Keita/DeeCLIP for research purposes.
MAAT: Mamba Adaptive Anomaly Transformer with association discrepancy for time series
Feb 11, 2025Abstract:Anomaly detection in time series is essential for industrial monitoring and environmental sensing, yet distinguishing anomalies from complex patterns remains challenging. Existing methods like the Anomaly Transformer and DCdetector have progressed, but they face limitations such as sensitivity to short-term contexts and inefficiency in noisy, non-stationary environments. To overcome these issues, we introduce MAAT, an improved architecture that enhances association discrepancy modeling and reconstruction quality. MAAT features Sparse Attention, efficiently capturing long-range dependencies by focusing on relevant time steps, thereby reducing computational redundancy. Additionally, a Mamba-Selective State Space Model is incorporated into the reconstruction module, utilizing a skip connection and Gated Attention to improve anomaly localization and detection performance. Extensive experiments show that MAAT significantly outperforms previous methods, achieving better anomaly distinguishability and generalization across various time series applications, setting a new standard for unsupervised time series anomaly detection in real-world scenarios.
Some Applications envisaged for the new generation of communications networks 6G
Jan 23, 2025Abstract:Future applications such as intelligent vehicles, the Internet of Things and holographic telepresence are already highlighting the limits of existing fifth-generation (5G) mobile networks. These limitations relate to data throughput, latency, reliability, availability, processing, connection density and global coverage, whether terrestrial, submarine or space-based. To remedy this, research institutes have begun to look beyond IMT2020, and as a result, 6G should provide effective solutions to 5G shortcomings. 6G will offer high quality of service and energy efficiency to meet the demands of future applications that are unimaginable to most people. In this article, we present the future applications and services promised by 6G.
Electrostatic Force Regularization for Neural Structured Pruning
Nov 17, 2024



Abstract:The demand for deploying deep convolutional neural networks (DCNNs) on resource-constrained devices for real-time applications remains substantial. However, existing state-of-the-art structured pruning methods often involve intricate implementations, require modifications to the original network architectures, and necessitate an extensive fine-tuning phase. To overcome these challenges, we propose a novel method that, for the first time, incorporates the concepts of charge and electrostatic force from physics into the training process of DCNNs. The magnitude of this force is directly proportional to the product of the charges of the convolution filter and the source filter, and inversely proportional to the square of the distance between them. We applied this electrostatic-like force to the convolution filters, either attracting filters with opposite charges toward non-zero weights or repelling filters with like charges toward zero weights. Consequently, filters subject to repulsive forces have their weights reduced to zero, enabling their removal, while the attractive forces preserve filters with significant weights that retain information. Unlike conventional methods, our approach is straightforward to implement, does not require any architectural modifications, and simultaneously optimizes weights and ranks filter importance, all without the need for extensive fine-tuning. We validated the efficacy of our method on modern DCNN architectures using the MNIST, CIFAR, and ImageNet datasets, achieving competitive performance compared to existing structured pruning approaches.
Boosting Hyperspectral Image Classification with Gate-Shift-Fuse Mechanisms in a Novel CNN-Transformer Approach
Jun 20, 2024Abstract:During the process of classifying Hyperspectral Image (HSI), every pixel sample is categorized under a land-cover type. CNN-based techniques for HSI classification have notably advanced the field by their adept feature representation capabilities. However, acquiring deep features remains a challenge for these CNN-based methods. In contrast, transformer models are adept at extracting high-level semantic features, offering a complementary strength. This paper's main contribution is the introduction of an HSI classification model that includes two convolutional blocks, a Gate-Shift-Fuse (GSF) block and a transformer block. This model leverages the strengths of CNNs in local feature extraction and transformers in long-range context modelling. The GSF block is designed to strengthen the extraction of local and global spatial-spectral features. An effective attention mechanism module is also proposed to enhance the extraction of information from HSI cubes. The proposed method is evaluated on four well-known datasets (the Indian Pines, Pavia University, WHU-WHU-Hi-LongKou and WHU-Hi-HanChuan), demonstrating that the proposed framework achieves superior results compared to other models.
Boosting House Price Estimations with Multi-Head Gated Attention
May 13, 2024



Abstract:Evaluating house prices is crucial for various stakeholders, including homeowners, investors, and policymakers. However, traditional spatial interpolation methods have limitations in capturing the complex spatial relationships that affect property values. To address these challenges, we have developed a new method called Multi-Head Gated Attention for spatial interpolation. Our approach builds upon attention-based interpolation models and incorporates multiple attention heads and gating mechanisms to capture spatial dependencies and contextual information better. Importantly, our model produces embeddings that reduce the dimensionality of the data, enabling simpler models like linear regression to outperform complex ensembling models. We conducted extensive experiments to compare our model with baseline methods and the original attention-based interpolation model. The results show a significant improvement in the accuracy of house price predictions, validating the effectiveness of our approach. This research advances the field of spatial interpolation and provides a robust tool for more precise house price evaluation. Our GitHub repository.contains the data and code for all datasets, which are available for researchers and practitioners interested in replicating or building upon our work.
D-TrAttUnet: Toward Hybrid CNN-Transformer Architecture for Generic and Subtle Segmentation in Medical Images
May 07, 2024



Abstract:Over the past two decades, machine analysis of medical imaging has advanced rapidly, opening up significant potential for several important medical applications. As complicated diseases increase and the number of cases rises, the role of machine-based imaging analysis has become indispensable. It serves as both a tool and an assistant to medical experts, providing valuable insights and guidance. A particularly challenging task in this area is lesion segmentation, a task that is challenging even for experienced radiologists. The complexity of this task highlights the urgent need for robust machine learning approaches to support medical staff. In response, we present our novel solution: the D-TrAttUnet architecture. This framework is based on the observation that different diseases often target specific organs. Our architecture includes an encoder-decoder structure with a composite Transformer-CNN encoder and dual decoders. The encoder includes two paths: the Transformer path and the Encoders Fusion Module path. The Dual-Decoder configuration uses two identical decoders, each with attention gates. This allows the model to simultaneously segment lesions and organs and integrate their segmentation losses. To validate our approach, we performed evaluations on the Covid-19 and Bone Metastasis segmentation tasks. We also investigated the adaptability of the model by testing it without the second decoder in the segmentation of glands and nuclei. The results confirmed the superiority of our approach, especially in Covid-19 infections and the segmentation of bone metastases. In addition, the hybrid encoder showed exceptional performance in the segmentation of glands and nuclei, solidifying its role in modern medical image analysis.
Artificial Intelligence in Bone Metastasis Analysis: Current Advancements, Opportunities and Challenges
Apr 30, 2024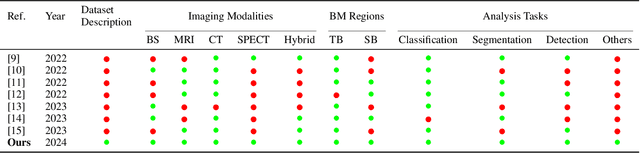
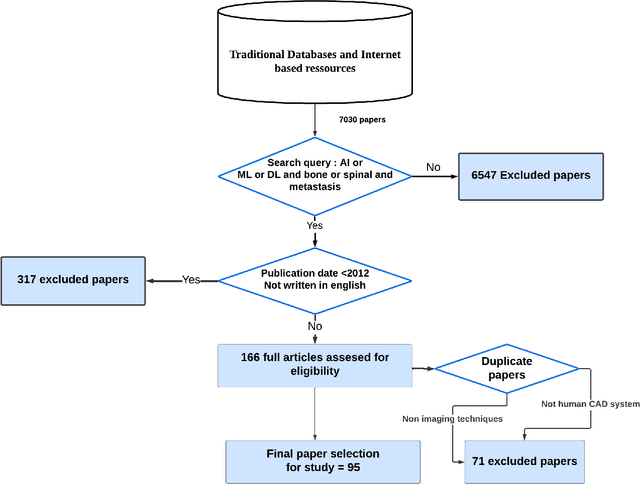
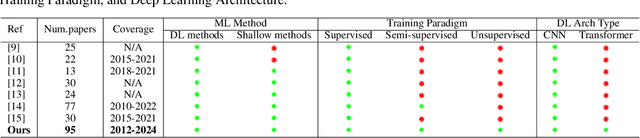
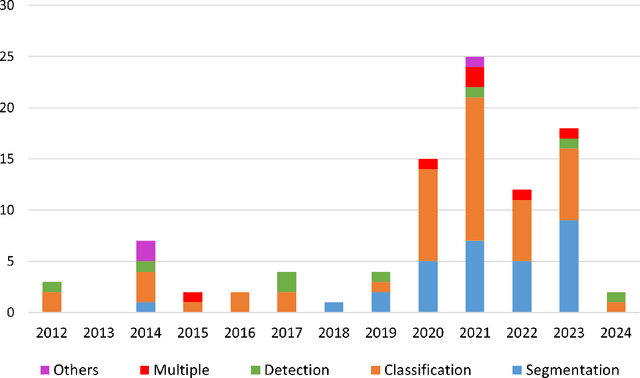
Abstract:In recent years, Artificial Intelligence (AI) has been widely used in medicine, particularly in the analysis of medical imaging, which has been driven by advances in computer vision and deep learning methods. This is particularly important in overcoming the challenges posed by diseases such as Bone Metastases (BM), a common and complex malignancy of the bones. Indeed, there have been an increasing interest in developing Machine Learning (ML) techniques into oncologic imaging for BM analysis. In order to provide a comprehensive overview of the current state-of-the-art and advancements for BM analysis using artificial intelligence, this review is conducted with the accordance with PRISMA guidelines. Firstly, this review highlights the clinical and oncologic perspectives of BM and the used medical imaging modalities, with discussing their advantages and limitations. Then the review focuses on modern approaches with considering the main BM analysis tasks, which includes: classification, detection and segmentation. The results analysis show that ML technologies can achieve promising performance for BM analysis and have significant potential to improve clinician efficiency and cope with time and cost limitations. Furthermore, there are requirements for further research to validate the clinical performance of ML tools and facilitate their integration into routine clinical practice.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge