A. Stephen McGough
O-TALC: Steps Towards Combating Oversegmentation within Online Action Segmentation
Apr 10, 2024Abstract:Online temporal action segmentation shows a strong potential to facilitate many HRI tasks where extended human action sequences must be tracked and understood in real time. Traditional action segmentation approaches, however, operate in an offline two stage approach, relying on computationally expensive video wide features for segmentation, rendering them unsuitable for online HRI applications. In order to facilitate online action segmentation on a stream of incoming video data, we introduce two methods for improved training and inference of backbone action recognition models, allowing them to be deployed directly for online frame level classification. Firstly, we introduce surround dense sampling whilst training to facilitate training vs. inference clip matching and improve segment boundary predictions. Secondly, we introduce an Online Temporally Aware Label Cleaning (O-TALC) strategy to explicitly reduce oversegmentation during online inference. As our methods are backbone invariant, they can be deployed with computationally efficient spatio-temporal action recognition models capable of operating in real time with a small segmentation latency. We show our method outperforms similar online action segmentation work as well as matches the performance of many offline models with access to full temporal resolution when operating on challenging fine-grained datasets.
Insights from the Use of Previously Unseen Neural Architecture Search Datasets
Apr 02, 2024Abstract:The boundless possibility of neural networks which can be used to solve a problem -- each with different performance -- leads to a situation where a Deep Learning expert is required to identify the best neural network. This goes against the hope of removing the need for experts. Neural Architecture Search (NAS) offers a solution to this by automatically identifying the best architecture. However, to date, NAS work has focused on a small set of datasets which we argue are not representative of real-world problems. We introduce eight new datasets created for a series of NAS Challenges: AddNIST, Language, MultNIST, CIFARTile, Gutenberg, Isabella, GeoClassing, and Chesseract. These datasets and challenges are developed to direct attention to issues in NAS development and to encourage authors to consider how their models will perform on datasets unknown to them at development time. We present experimentation using standard Deep Learning methods as well as the best results from challenge participants.
The Effects of Signal-to-Noise Ratio on Generative Adversarial Networks Applied to Marine Bioacoustic Data
Dec 22, 2023Abstract:In recent years generative adversarial networks (GANs) have been used to supplement datasets within the field of marine bioacoustics. This is driven by factors such as the cost to collect data, data sparsity and aid preprocessing. One notable challenge with marine bioacoustic data is the low signal-to-noise ratio (SNR) posing difficulty when applying deep learning techniques such as GANs. This work investigates the effect SNR has on the audio-based GAN performance and examines three different evaluation methodologies for GAN performance, yielding interesting results on the effects of SNR on GANs, specifically WaveGAN.
The Forecastability of Underlying Building Electricity Demand from Time Series Data
Nov 29, 2023



Abstract:Forecasting building energy consumption has become a promising solution in Building Energy Management Systems for energy saving and optimization. Furthermore, it can play an important role in the efficient management of the operation of a smart grid. Different data-driven approaches to forecast the future energy demand of buildings at different scale, and over various time horizons, can be found in the scientific literature, including extensive Machine Learning and Deep Learning approaches. However, the identification of the most accurate forecaster model which can be utilized to predict the energy demand of such a building is still challenging.In this paper, the design and implementation of a data-driven approach to predict how forecastable the future energy demand of a building is, without first utilizing a data-driven forecasting model, is presented. The investigation utilizes a historical electricity consumption time series data set with a half-hour interval that has been collected from a group of residential buildings located in the City of London, United Kingdom
How much data do I need? A case study on medical data
Nov 26, 2023Abstract:The collection of data to train a Deep Learning network is costly in terms of effort and resources. In many cases, especially in a medical context, it may have detrimental impacts. Such as requiring invasive medical procedures or processes which could in themselves cause medical harm. However, Deep Learning is seen as a data hungry method. Here, we look at two commonly held adages i) more data gives better results and ii) transfer learning will aid you when you don't have enough data. These are widely assumed to be true and used as evidence for choosing how to solve a problem when Deep Learning is involved. We evaluate six medical datasets and six general datasets. Training a ResNet18 network on varying subsets of these datasets to evaluate `more data gives better results'. We take eleven of these datasets as the sources for Transfer Learning on subsets of the twelfth dataset -- Chest -- in order to determine whether Transfer Learning is universally beneficial. We go further to see whether multi-stage Transfer Learning provides a consistent benefit. Our analysis shows that the real situation is more complex than these simple adages -- more data could lead to a case of diminishing returns and an incorrect choice of dataset for transfer learning can lead to worse performance, with datasets which we would consider highly similar to the Chest dataset giving worse results than datasets which are more dissimilar. Multi-stage transfer learning likewise reveals complex relationships between datasets.
NCL-SM: A Fully Annotated Dataset of Images from Human Skeletal Muscle Biopsies
Nov 25, 2023



Abstract:Single cell analysis of human skeletal muscle (SM) tissue cross-sections is a fundamental tool for understanding many neuromuscular disorders. For this analysis to be reliable and reproducible, identification of individual fibres within microscopy images (segmentation) of SM tissue should be automatic and precise. Biomedical scientists in this field currently rely on custom tools and general machine learning (ML) models, both followed by labour intensive and subjective manual interventions to fine-tune segmentation. We believe that fully automated, precise, reproducible segmentation is possible by training ML models. However, in this important biomedical domain, there are currently no good quality, publicly available annotated imaging datasets available for ML model training. In this paper we release NCL-SM: a high quality bioimaging dataset of 46 human SM tissue cross-sections from both healthy control subjects and from patients with genetically diagnosed muscle pathology. These images include $>$ 50k manually segmented muscle fibres (myofibres). In addition we also curated high quality myofibre segmentations, annotating reasons for rejecting low quality myofibres and low quality regions in SM tissue images, making these annotations completely ready for downstream analysis. This, we believe, will pave the way for development of a fully automatic pipeline that identifies individual myofibres within images of tissue sections and, in particular, also classifies individual myofibres that are fit for further analysis.
Introducing NCL-SM: A Fully Annotated Dataset of Images from Human Skeletal Muscle Biopsies
Nov 18, 2023


Abstract:Single cell analysis of skeletal muscle (SM) tissue is a fundamental tool for understanding many neuromuscular disorders. For this analysis to be reliable and reproducible, identification of individual fibres within microscopy images (segmentation) of SM tissue should be precise. There is currently no tool or pipeline that makes automatic and precise segmentation and curation of images of SM tissue cross-sections possible. Biomedical scientists in this field rely on custom tools and general machine learning (ML) models, both followed by labour intensive and subjective manual interventions to get the segmentation right. We believe that automated, precise, reproducible segmentation is possible by training ML models. However, there are currently no good quality, publicly available annotated imaging datasets available for ML model training. In this paper we release NCL-SM: a high quality bioimaging dataset of 46 human tissue sections from healthy control subjects and from patients with genetically diagnosed muscle pathology. These images include $>$ 50k manually segmented muscle fibres (myofibres). In addition we also curated high quality myofibres and annotated reasons for rejecting low quality myofibres and regions in SM tissue images, making this data completely ready for downstream analysis. This, we believe, will pave the way for development of a fully automatic pipeline that identifies individual myofibres within images of tissue sections and, in particular, also classifies individual myofibres that are fit for further analysis.
Towards Automatic Cetacean Photo-Identification: A Framework for Fine-Grain, Few-Shot Learning in Marine Ecology
Dec 07, 2022



Abstract:Photo-identification (photo-id) is one of the main non-invasive capture-recapture methods utilised by marine researchers for monitoring cetacean (dolphin, whale, and porpoise) populations. This method has historically been performed manually resulting in high workload and cost due to the vast number of images collected. Recently automated aids have been developed to help speed-up photo-id, although they are often disjoint in their processing and do not utilise all available identifying information. Work presented in this paper aims to create a fully automatic photo-id aid capable of providing most likely matches based on all available information without the need for data pre-processing such as cropping. This is achieved through a pipeline of computer vision models and post-processing techniques aimed at detecting cetaceans in unedited field imagery before passing them downstream for individual level catalogue matching. The system is capable of handling previously uncatalogued individuals and flagging these for investigation thanks to catalogue similarity comparison. We evaluate the system against multiple real-life photo-id catalogues, achieving mAP@IOU[0.5] = 0.91, 0.96 for the task of dorsal fin detection on catalogues from Tanzania and the UK respectively and 83.1, 97.5% top-10 accuracy for the task of individual classification on catalogues from the UK and USA.
Explainable Deep Learning to Profile Mitochondrial Disease Using High Dimensional Protein Expression Data
Oct 31, 2022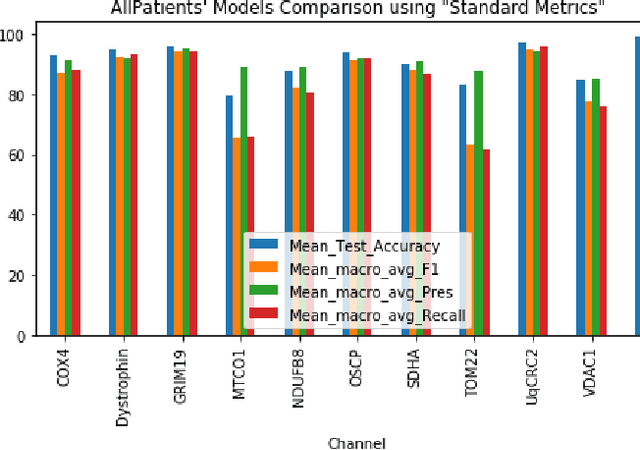
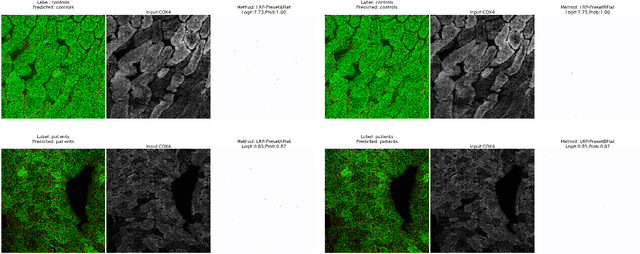
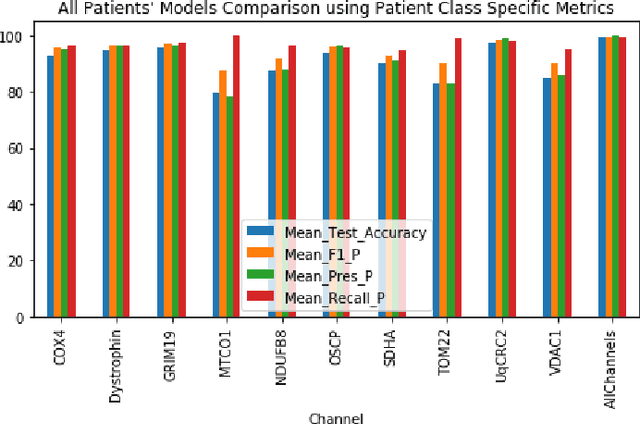
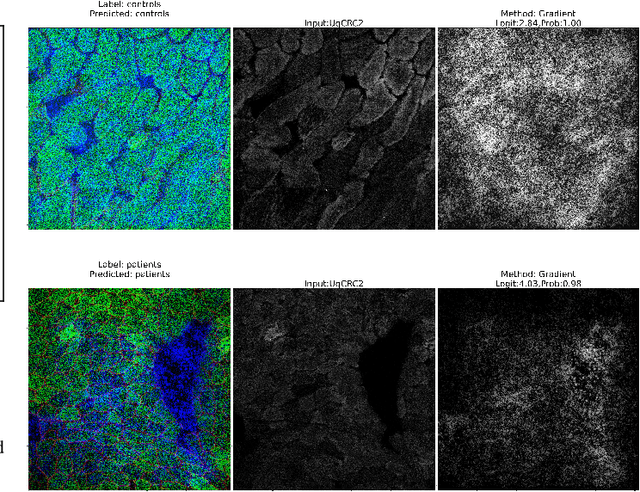
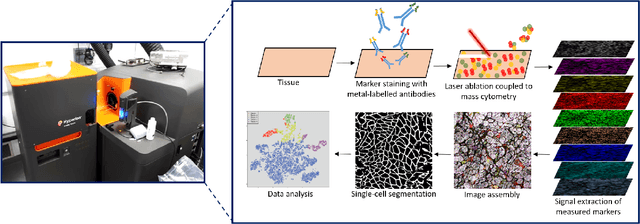
Abstract:Mitochondrial diseases are currently untreatable due to our limited understanding of their pathology. We study the expression of various mitochondrial proteins in skeletal myofibres (SM) in order to discover processes involved in mitochondrial pathology using Imaging Mass Cytometry (IMC). IMC produces high dimensional multichannel pseudo-images representing spatial variation in the expression of a panel of proteins within a tissue, including subcellular variation. Statistical analysis of these images requires semi-automated annotation of thousands of SMs in IMC images of patient muscle biopsies. In this paper we investigate the use of deep learning (DL) on raw IMC data to analyse it without any manual pre-processing steps, statistical summaries or statistical models. For this we first train state-of-art computer vision DL models on all available image channels, both combined and individually. We observed better than expected accuracy for many of these models. We then apply state-of-the-art explainable techniques relevant to computer vision DL to find the basis of the predictions of these models. Some of the resulting visual explainable maps highlight features in the images that appear consistent with the latest hypotheses about mitochondrial disease progression within myofibres.
Optimizing a domestic battery and solar photovoltaic system with deep reinforcement learning
Sep 10, 2021
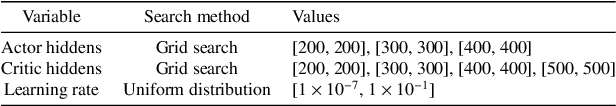
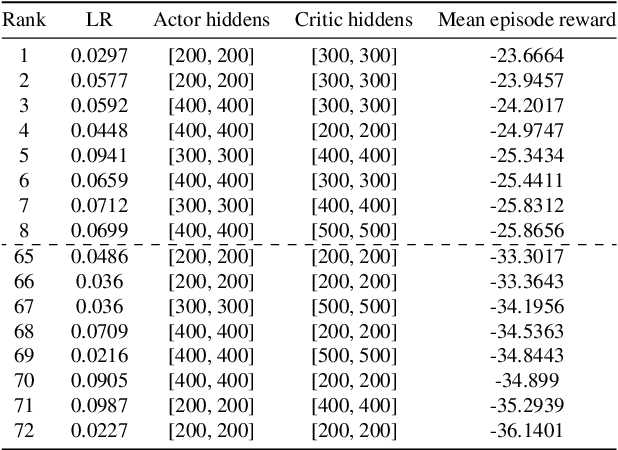
Abstract:A lowering in the cost of batteries and solar PV systems has led to a high uptake of solar battery home systems. In this work, we use the deep deterministic policy gradient algorithm to optimise the charging and discharging behaviour of a battery within such a system. Our approach outputs a continuous action space when it charges and discharges the battery, and can function well in a stochastic environment. We show good performance of this algorithm by lowering the expenditure of a single household on electricity to almost \$1AUD for large batteries across selected weeks within a year.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge