Atif Khan
NCL-SM: A Fully Annotated Dataset of Images from Human Skeletal Muscle Biopsies
Nov 25, 2023



Abstract:Single cell analysis of human skeletal muscle (SM) tissue cross-sections is a fundamental tool for understanding many neuromuscular disorders. For this analysis to be reliable and reproducible, identification of individual fibres within microscopy images (segmentation) of SM tissue should be automatic and precise. Biomedical scientists in this field currently rely on custom tools and general machine learning (ML) models, both followed by labour intensive and subjective manual interventions to fine-tune segmentation. We believe that fully automated, precise, reproducible segmentation is possible by training ML models. However, in this important biomedical domain, there are currently no good quality, publicly available annotated imaging datasets available for ML model training. In this paper we release NCL-SM: a high quality bioimaging dataset of 46 human SM tissue cross-sections from both healthy control subjects and from patients with genetically diagnosed muscle pathology. These images include $>$ 50k manually segmented muscle fibres (myofibres). In addition we also curated high quality myofibre segmentations, annotating reasons for rejecting low quality myofibres and low quality regions in SM tissue images, making these annotations completely ready for downstream analysis. This, we believe, will pave the way for development of a fully automatic pipeline that identifies individual myofibres within images of tissue sections and, in particular, also classifies individual myofibres that are fit for further analysis.
Introducing NCL-SM: A Fully Annotated Dataset of Images from Human Skeletal Muscle Biopsies
Nov 18, 2023


Abstract:Single cell analysis of skeletal muscle (SM) tissue is a fundamental tool for understanding many neuromuscular disorders. For this analysis to be reliable and reproducible, identification of individual fibres within microscopy images (segmentation) of SM tissue should be precise. There is currently no tool or pipeline that makes automatic and precise segmentation and curation of images of SM tissue cross-sections possible. Biomedical scientists in this field rely on custom tools and general machine learning (ML) models, both followed by labour intensive and subjective manual interventions to get the segmentation right. We believe that automated, precise, reproducible segmentation is possible by training ML models. However, there are currently no good quality, publicly available annotated imaging datasets available for ML model training. In this paper we release NCL-SM: a high quality bioimaging dataset of 46 human tissue sections from healthy control subjects and from patients with genetically diagnosed muscle pathology. These images include $>$ 50k manually segmented muscle fibres (myofibres). In addition we also curated high quality myofibres and annotated reasons for rejecting low quality myofibres and regions in SM tissue images, making this data completely ready for downstream analysis. This, we believe, will pave the way for development of a fully automatic pipeline that identifies individual myofibres within images of tissue sections and, in particular, also classifies individual myofibres that are fit for further analysis.
Deep Ensembling for Perceptual Image Quality Assessment
May 16, 2023Abstract:Blind image quality assessment is a challenging task particularly due to the unavailability of reference information. Training a deep neural network requires a large amount of training data which is not readily available for image quality. Transfer learning is usually opted to overcome this limitation and different deep architectures are used for this purpose as they learn features differently. After extensive experiments, we have designed a deep architecture containing two CNN architectures as its sub-units. Moreover, a self-collected image database BIQ2021 is proposed with 12,000 images having natural distortions. The self-collected database is subjectively scored and is used for model training and validation. It is demonstrated that synthetic distortion databases cannot provide generalization beyond the distortion types used in the database and they are not ideal candidates for general-purpose image quality assessment. Moreover, a large-scale database of 18.75 million images with synthetic distortions is used to pretrain the model and then retrain it on benchmark databases for evaluation. Experiments are conducted on six benchmark databases three of which are synthetic distortion databases (LIVE, CSIQ and TID2013) and three are natural distortion databases (LIVE Challenge Database, CID2013 and KonIQ-10 k). The proposed approach has provided a Pearson correlation coefficient of 0.8992, 0.8472 and 0.9452 subsequently and Spearman correlation coefficient of 0.8863, 0.8408 and 0.9421. Moreover, the performance is demonstrated using perceptually weighted rank correlation to indicate the perceptual superiority of the proposed approach. Multiple experiments are conducted to validate the generalization performance of the proposed model by training on different subsets of the databases and validating on the test subset of BIQ2021 database.
Explainable Deep Learning to Profile Mitochondrial Disease Using High Dimensional Protein Expression Data
Oct 31, 2022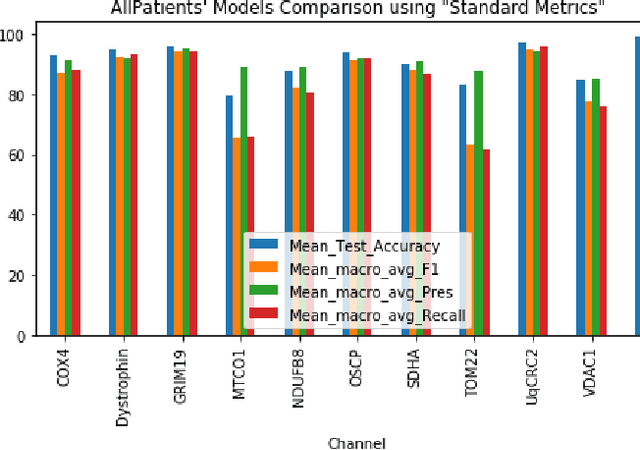
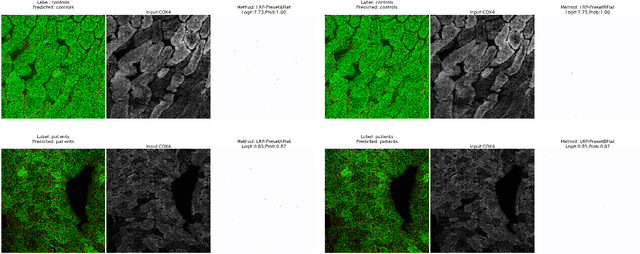
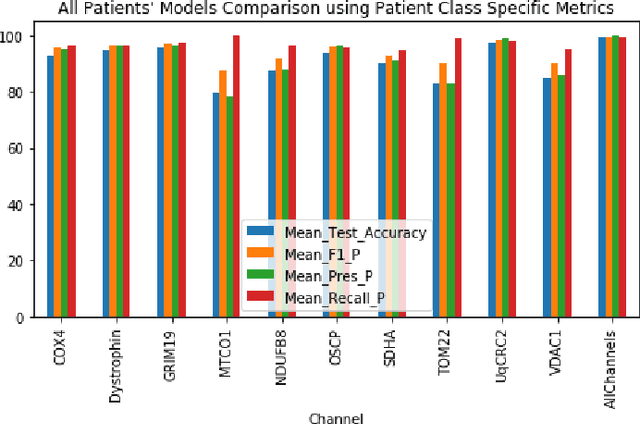
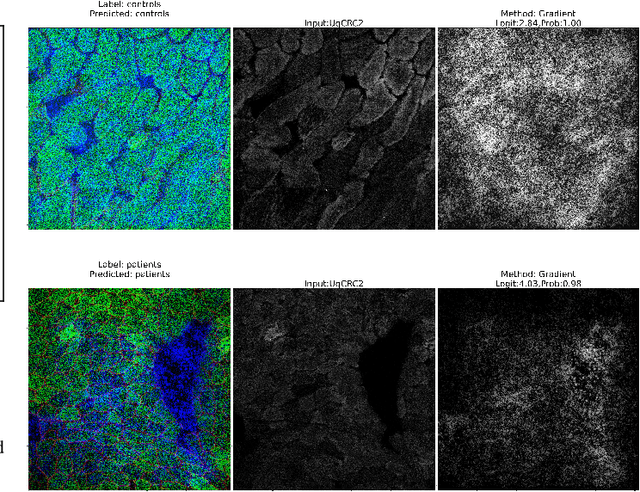
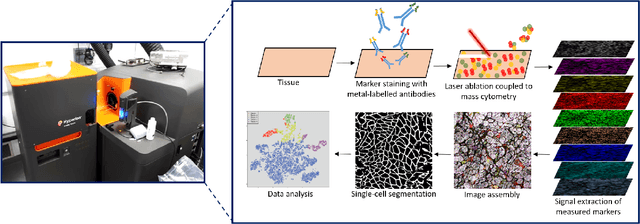
Abstract:Mitochondrial diseases are currently untreatable due to our limited understanding of their pathology. We study the expression of various mitochondrial proteins in skeletal myofibres (SM) in order to discover processes involved in mitochondrial pathology using Imaging Mass Cytometry (IMC). IMC produces high dimensional multichannel pseudo-images representing spatial variation in the expression of a panel of proteins within a tissue, including subcellular variation. Statistical analysis of these images requires semi-automated annotation of thousands of SMs in IMC images of patient muscle biopsies. In this paper we investigate the use of deep learning (DL) on raw IMC data to analyse it without any manual pre-processing steps, statistical summaries or statistical models. For this we first train state-of-art computer vision DL models on all available image channels, both combined and individually. We observed better than expected accuracy for many of these models. We then apply state-of-the-art explainable techniques relevant to computer vision DL to find the basis of the predictions of these models. Some of the resulting visual explainable maps highlight features in the images that appear consistent with the latest hypotheses about mitochondrial disease progression within myofibres.
Lightweight Mobile Automated Assistant-to-physician for Global Lower-resource Areas
Oct 28, 2021Abstract:Importance: Lower-resource areas in Africa and Asia face a unique set of healthcare challenges: the dual high burden of communicable and non-communicable diseases; a paucity of highly trained primary healthcare providers in both rural and densely populated urban areas; and a lack of reliable, inexpensive internet connections. Objective: To address these challenges, we designed an artificial intelligence assistant to help primary healthcare providers in lower-resource areas document demographic and medical sign/symptom data and to record and share diagnostic data in real-time with a centralized database. Design: We trained our system using multiple data sets, including US-based electronic medical records (EMRs) and open-source medical literature and developed an adaptive, general medical assistant system based on machine learning algorithms. Main outcomes and Measure: The application collects basic information from patients and provides primary care providers with diagnoses and prescriptions suggestions. The application is unique from existing systems in that it covers a wide range of common diseases, signs, and medication typical in lower-resource countries; the application works with or without an active internet connection. Results: We have built and implemented an adaptive learning system that assists trained primary care professionals by means of an Android smartphone application, which interacts with a central database and collects real-time data. The application has been tested by dozens of primary care providers. Conclusions and Relevance: Our application would provide primary healthcare providers in lower-resource areas with a tool that enables faster and more accurate documentation of medical encounters. This application could be leveraged to automatically populate local or national EMR systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge