Zhaoguo Wang
Let It Flow: Agentic Crafting on Rock and Roll, Building the ROME Model within an Open Agentic Learning Ecosystem
Dec 31, 2025Abstract:Agentic crafting requires LLMs to operate in real-world environments over multiple turns by taking actions, observing outcomes, and iteratively refining artifacts. Despite its importance, the open-source community lacks a principled, end-to-end ecosystem to streamline agent development. We introduce the Agentic Learning Ecosystem (ALE), a foundational infrastructure that optimizes the production pipeline for agent LLMs. ALE consists of three components: ROLL, a post-training framework for weight optimization; ROCK, a sandbox environment manager for trajectory generation; and iFlow CLI, an agent framework for efficient context engineering. We release ROME (ROME is Obviously an Agentic Model), an open-source agent grounded by ALE and trained on over one million trajectories. Our approach includes data composition protocols for synthesizing complex behaviors and a novel policy optimization algorithm, Interaction-based Policy Alignment (IPA), which assigns credit over semantic interaction chunks rather than individual tokens to improve long-horizon training stability. Empirically, we evaluate ROME within a structured setting and introduce Terminal Bench Pro, a benchmark with improved scale and contamination control. ROME demonstrates strong performance across benchmarks like SWE-bench Verified and Terminal Bench, proving the effectiveness of the ALE infrastructure.
An AI-native experimental laboratory for autonomous biomolecular engineering
Jul 03, 2025



Abstract:Autonomous scientific research, capable of independently conducting complex experiments and serving non-specialists, represents a long-held aspiration. Achieving it requires a fundamental paradigm shift driven by artificial intelligence (AI). While autonomous experimental systems are emerging, they remain confined to areas featuring singular objectives and well-defined, simple experimental workflows, such as chemical synthesis and catalysis. We present an AI-native autonomous laboratory, targeting highly complex scientific experiments for applications like autonomous biomolecular engineering. This system autonomously manages instrumentation, formulates experiment-specific procedures and optimization heuristics, and concurrently serves multiple user requests. Founded on a co-design philosophy of models, experiments, and instruments, the platform supports the co-evolution of AI models and the automation system. This establishes an end-to-end, multi-user autonomous laboratory that handles complex, multi-objective experiments across diverse instrumentation. Our autonomous laboratory supports fundamental nucleic acid functions-including synthesis, transcription, amplification, and sequencing. It also enables applications in fields such as disease diagnostics, drug development, and information storage. Without human intervention, it autonomously optimizes experimental performance to match state-of-the-art results achieved by human scientists. In multi-user scenarios, the platform significantly improves instrument utilization and experimental efficiency. This platform paves the way for advanced biomaterials research to overcome dependencies on experts and resource barriers, establishing a blueprint for science-as-a-service at scale.
Accelerating Large Language Models through Partially Linear Feed-Forward Network
Jan 17, 2025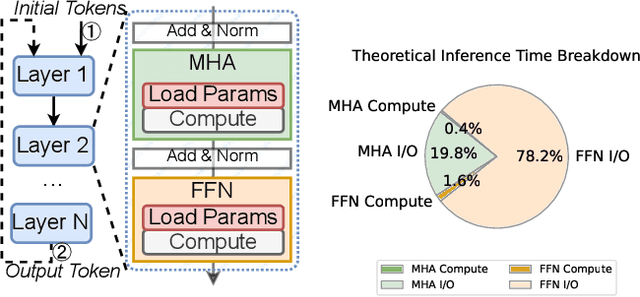
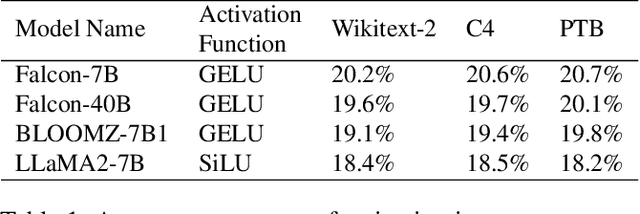
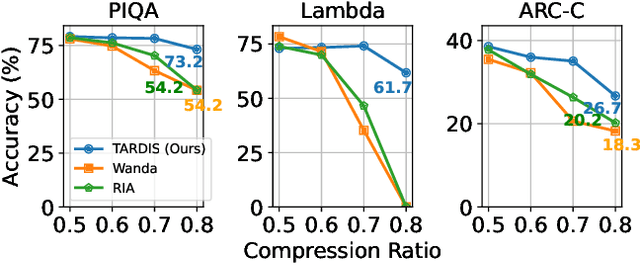
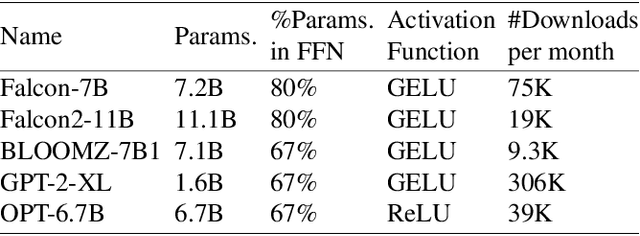
Abstract:Large language models (LLMs) demonstrate remarkable capabilities but face deployment challenges due to their massive parameter counts. While existing compression techniques like pruning can reduce model size, it leads to significant accuracy degradation under high compression ratios. We present a novel perspective inspired by constant folding in compiler optimization. Our approach enables parameter reduction by treating activation functions in LLMs as linear functions. However, recent LLMs use complex non-linear activations like GELU that prevent direct application of this technique. We propose TARDIS, which enables optimization of LLMs with non-linear activations by partially approximating them with linear functions in frequently occurring input ranges. For outlier inputs, TARDIS employs an online predictor to dynamically fall back to original computations. Our experiments demonstrate that TARDIS achieves 80% parameter reduction in feed-forward networks, while significantly outperforming state-of-the-art pruning methods Wanda and RIA with up to 65% higher accuracy. In practical deployments for a 7B model, TARDIS achieves 1.6x end-to-end inference speedup when integrated with the vLLM serving system, and 1.4x speedup with the widely adopted HuggingFace implementation, while incurring only a 10.9% accuracy trade-off.
Learned Indexes for Dynamic Workloads
Feb 02, 2019


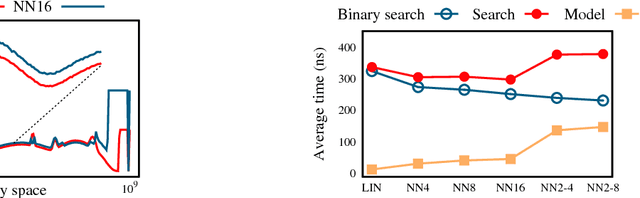
Abstract:The recent proposal of learned index structures opens up a new perspective on how traditional range indexes can be optimized. However, the current learned indexes assume the data distribution is relatively static and the access pattern is uniform, while real-world scenarios consist of skew query distribution and evolving data. In this paper, we demonstrate that the missing consideration of access patterns and dynamic data distribution notably hinders the applicability of learned indexes. To this end, we propose solutions for learned indexes for dynamic workloads (called Doraemon). To improve the latency for skew queries, Doraemon augments the training data with access frequencies. To address the slow model re-training when data distribution shifts, Doraemon caches the previously-trained models and incrementally fine-tunes them for similar access patterns and data distribution. Our preliminary result shows that, Doraemon improves the query latency by 45.1% and reduces the model re-training time to 1/20.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge