Yusung Kim
Graph-Assisted Stitching for Offline Hierarchical Reinforcement Learning
Jun 09, 2025Abstract:Existing offline hierarchical reinforcement learning methods rely on high-level policy learning to generate subgoal sequences. However, their efficiency degrades as task horizons increase, and they lack effective strategies for stitching useful state transitions across different trajectories. We propose Graph-Assisted Stitching (GAS), a novel framework that formulates subgoal selection as a graph search problem rather than learning an explicit high-level policy. By embedding states into a Temporal Distance Representation (TDR) space, GAS clusters semantically similar states from different trajectories into unified graph nodes, enabling efficient transition stitching. A shortest-path algorithm is then applied to select subgoal sequences within the graph, while a low-level policy learns to reach the subgoals. To improve graph quality, we introduce the Temporal Efficiency (TE) metric, which filters out noisy or inefficient transition states, significantly enhancing task performance. GAS outperforms prior offline HRL methods across locomotion, navigation, and manipulation tasks. Notably, in the most stitching-critical task, it achieves a score of 88.3, dramatically surpassing the previous state-of-the-art score of 1.0. Our source code is available at: https://github.com/qortmdgh4141/GAS.
Self-Predictive Dynamics for Generalization of Vision-based Reinforcement Learning
Jun 05, 2025Abstract:Vision-based reinforcement learning requires efficient and robust representations of image-based observations, especially when the images contain distracting (task-irrelevant) elements such as shadows, clouds, and light. It becomes more important if those distractions are not exposed during training. We design a Self-Predictive Dynamics (SPD) method to extract task-relevant features efficiently, even in unseen observations after training. SPD uses weak and strong augmentations in parallel, and learns representations by predicting inverse and forward transitions across the two-way augmented versions. In a set of MuJoCo visual control tasks and an autonomous driving task (CARLA), SPD outperforms previous studies in complex observations, and significantly improves the generalization performance for unseen observations. Our code is available at https://github.com/unigary/SPD.
Self-supervised One-Stage Learning for RF-based Multi-Person Pose Estimation
Jun 05, 2025Abstract:In the field of Multi-Person Pose Estimation (MPPE), Radio Frequency (RF)-based methods can operate effectively regardless of lighting conditions and obscured line-of-sight situations. Existing RF-based MPPE methods typically involve either 1) converting RF signals into heatmap images through complex preprocessing, or 2) applying a deep embedding network directly to raw RF signals. The first approach, while delivering decent performance, is computationally intensive and time-consuming. The second method, though simpler in preprocessing, results in lower MPPE accuracy and generalization performance. This paper proposes an efficient and lightweight one-stage MPPE model based on raw RF signals. By sub-grouping RF signals and embedding them using a shared single-layer CNN followed by multi-head attention, this model outperforms previous methods that embed all signals at once through a large and deep CNN. Additionally, we propose a new self-supervised learning (SSL) method that takes inputs from both one unmasked subgroup and the remaining masked subgroups to predict the latent representations of the masked data. Empirical results demonstrate that our model improves MPPE accuracy by up to 15 in PCKh@0.5 compared to previous methods using raw RF signals. Especially, the proposed SSL method has shown to significantly enhance performance improvements when placed in new locations or in front of obstacles at RF antennas, contributing to greater performance gains as the number of people increases. Our code and dataset is open at Github. https://github.com/sshnan7/SOSPE .
Dream to Generalize: Zero-Shot Model-Based Reinforcement Learning for Unseen Visual Distractions
Jun 05, 2025



Abstract:Model-based reinforcement learning (MBRL) has been used to efficiently solve vision-based control tasks in highdimensional image observations. Although recent MBRL algorithms perform well in trained observations, they fail when faced with visual distractions in observations. These task-irrelevant distractions (e.g., clouds, shadows, and light) may be constantly present in real-world scenarios. In this study, we propose a novel self-supervised method, Dream to Generalize (Dr. G), for zero-shot MBRL. Dr. G trains its encoder and world model with dual contrastive learning which efficiently captures task-relevant features among multi-view data augmentations. We also introduce a recurrent state inverse dynamics model that helps the world model to better understand the temporal structure. The proposed methods can enhance the robustness of the world model against visual distractions. To evaluate the generalization performance, we first train Dr. G on simple backgrounds and then test it on complex natural video backgrounds in the DeepMind Control suite, and the randomizing environments in Robosuite. Dr. G yields a performance improvement of 117% and 14% over prior works, respectively. Our code is open-sourced and available at https://github.com/JeongsooHa/DrG.git
Cross-Domain Semantic Segmentation on Inconsistent Taxonomy using VLMs
Aug 05, 2024



Abstract:The challenge of semantic segmentation in Unsupervised Domain Adaptation (UDA) emerges not only from domain shifts between source and target images but also from discrepancies in class taxonomies across domains. Traditional UDA research assumes consistent taxonomy between the source and target domains, thereby limiting their ability to recognize and adapt to the taxonomy of the target domain. This paper introduces a novel approach, Cross-Domain Semantic Segmentation on Inconsistent Taxonomy using Vision Language Models (CSI), which effectively performs domain-adaptive semantic segmentation even in situations of source-target class mismatches. CSI leverages the semantic generalization potential of Visual Language Models (VLMs) to create synergy with previous UDA methods. It leverages segment reasoning obtained through traditional UDA methods, combined with the rich semantic knowledge embedded in VLMs, to relabel new classes in the target domain. This approach allows for effective adaptation to extended taxonomies without requiring any ground truth label for the target domain. Our method has shown to be effective across various benchmarks in situations of inconsistent taxonomy settings (coarse-to-fine taxonomy and open taxonomy) and demonstrates consistent synergy effects when integrated with previous state-of-the-art UDA methods. The implementation is available at http://github.com/jkee58/CSI.
Federated Learning Enables Big Data for Rare Cancer Boundary Detection
Apr 25, 2022Abstract:Although machine learning (ML) has shown promise in numerous domains, there are concerns about generalizability to out-of-sample data. This is currently addressed by centrally sharing ample, and importantly diverse, data from multiple sites. However, such centralization is challenging to scale (or even not feasible) due to various limitations. Federated ML (FL) provides an alternative to train accurate and generalizable ML models, by only sharing numerical model updates. Here we present findings from the largest FL study to-date, involving data from 71 healthcare institutions across 6 continents, to generate an automatic tumor boundary detector for the rare disease of glioblastoma, utilizing the largest dataset of such patients ever used in the literature (25,256 MRI scans from 6,314 patients). We demonstrate a 33% improvement over a publicly trained model to delineate the surgically targetable tumor, and 23% improvement over the tumor's entire extent. We anticipate our study to: 1) enable more studies in healthcare informed by large and diverse data, ensuring meaningful results for rare diseases and underrepresented populations, 2) facilitate further quantitative analyses for glioblastoma via performance optimization of our consensus model for eventual public release, and 3) demonstrate the effectiveness of FL at such scale and task complexity as a paradigm shift for multi-site collaborations, alleviating the need for data sharing.
What does AI see? Deep segmentation networks discover biomarkers for lung cancer survival
Mar 26, 2019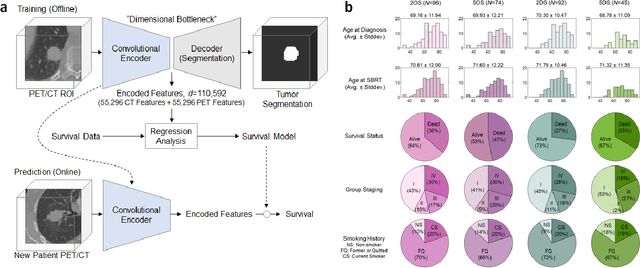
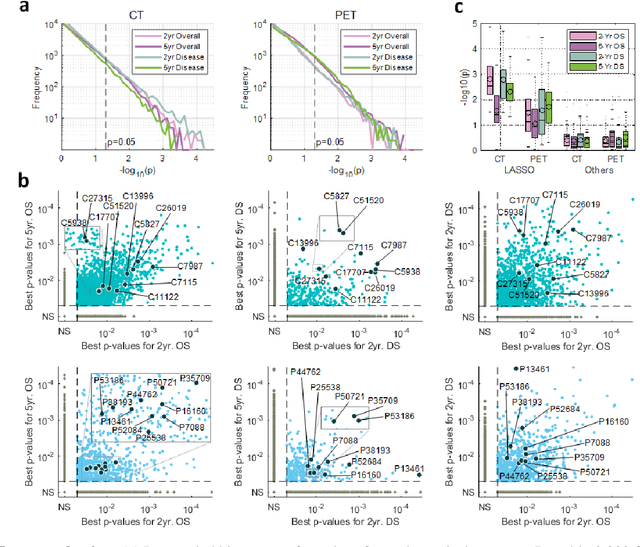
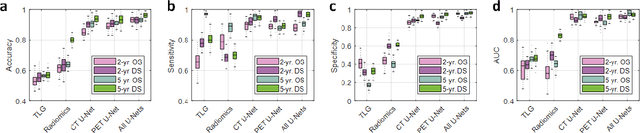
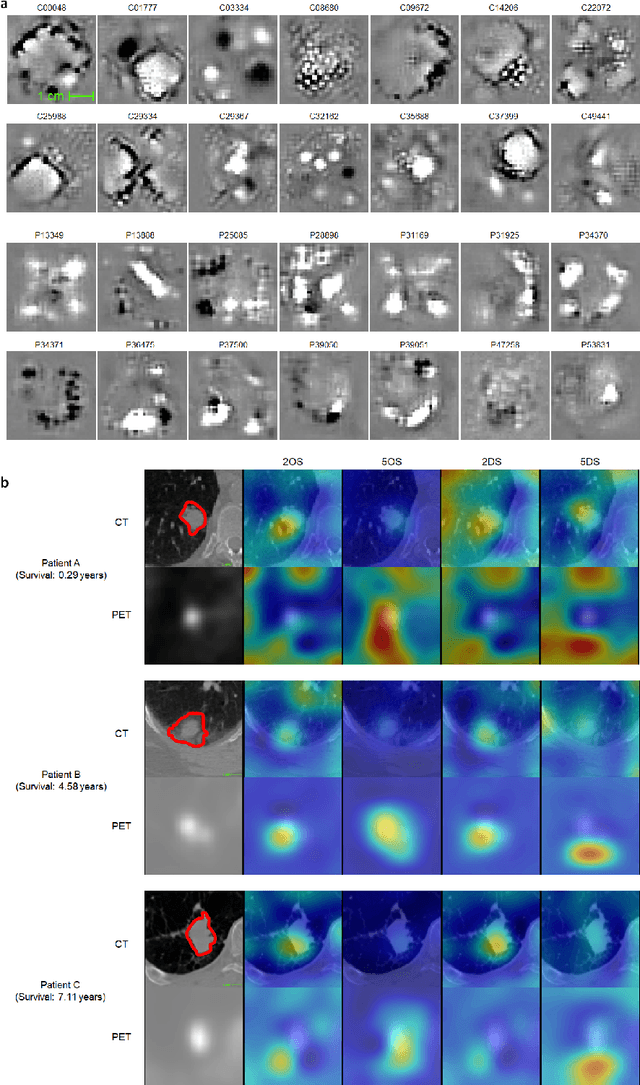
Abstract:Non-small-cell lung cancer (NSCLC) represents approximately 80-85% of lung cancer diagnoses and is the leading cause of cancer-related death worldwide. Recent studies indicate that image-based radiomics features from positron emission tomography-computed tomography (PET/CT) images have predictive power on NSCLC outcomes. To this end, easily calculated functional features such as the maximum and the mean of standard uptake value (SUV) and total lesion glycolysis (TLG) are most commonly used for NSCLC prognostication, but their prognostic value remains controversial. Meanwhile, convolutional neural networks (CNN) are rapidly emerging as a new premise for cancer image analysis, with significantly enhanced predictive power compared to other hand-crafted radiomics features. Here we show that CNN trained to perform the tumor segmentation task, with no other information than physician contours, identify a rich set of survival-related image features with remarkable prognostic value. In a retrospective study on 96 NSCLC patients before stereotactic-body radiotherapy (SBRT), we found that the CNN segmentation algorithm (U-Net) trained for tumor segmentation in PET/CT images, contained features having strong correlation with 2- and 5-year overall and disease-specific survivals. The U-net algorithm has not seen any other clinical information (e.g. survival, age, smoking history) than the images and the corresponding tumor contours provided by physicians. Furthermore, through visualization of the U-Net, we also found convincing evidence that the regions of progression appear to match with the regions where the U-Net features identified patterns that predicted higher likelihood of death. We anticipate our findings will be a starting point for more sophisticated non-intrusive patient specific cancer prognosis determination.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge