Yonghuang Wu
CCSD: Cross-Modal Compositional Self-Distillation for Robust Brain Tumor Segmentation with Missing Modalities
Nov 18, 2025



Abstract:The accurate segmentation of brain tumors from multi-modal MRI is critical for clinical diagnosis and treatment planning. While integrating complementary information from various MRI sequences is a common practice, the frequent absence of one or more modalities in real-world clinical settings poses a significant challenge, severely compromising the performance and generalizability of deep learning-based segmentation models. To address this challenge, we propose a novel Cross-Modal Compositional Self-Distillation (CCSD) framework that can flexibly handle arbitrary combinations of input modalities. CCSD adopts a shared-specific encoder-decoder architecture and incorporates two self-distillation strategies: (i) a hierarchical modality self-distillation mechanism that transfers knowledge across modality hierarchies to reduce semantic discrepancies, and (ii) a progressive modality combination distillation approach that enhances robustness to missing modalities by simulating gradual modality dropout during training. Extensive experiments on public brain tumor segmentation benchmarks demonstrate that CCSD achieves state-of-the-art performance across various missing-modality scenarios, with strong generalization and stability.
Instance-level Randomization: Toward More Stable LLM Evaluations
Sep 16, 2025



Abstract:Evaluations of large language models (LLMs) suffer from instability, where small changes of random factors such as few-shot examples can lead to drastic fluctuations of scores and even model rankings. Moreover, different LLMs can have different preferences for a certain setting of random factors. As a result, using a fixed setting of random factors, which is often adopted as the paradigm of current evaluations, can lead to potential unfair comparisons between LLMs. To mitigate the volatility of evaluations, we first theoretically analyze the sources of variance induced by changes in random factors. Targeting these specific sources, we then propose the instance-level randomization (ILR) method to reduce variance and enhance fairness in model comparisons. Instead of using a fixed setting across the whole benchmark in a single experiment, we randomize all factors that affect evaluation scores for every single instance, run multiple experiments and report the averaged score. Theoretical analyses and empirical results demonstrate that ILR can reduce the variance and unfair comparisons caused by random factors, as well as achieve similar robustness level with less than half computational cost compared with previous methods.
Cross-Patient Pseudo Bags Generation and Curriculum Contrastive Learning for Imbalanced Multiclassification of Whole Slide Image
Nov 18, 2024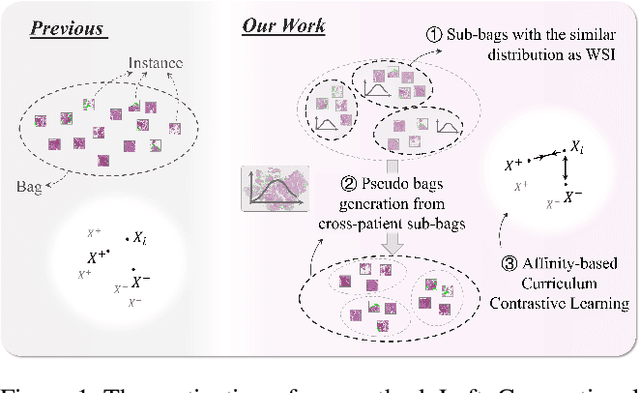
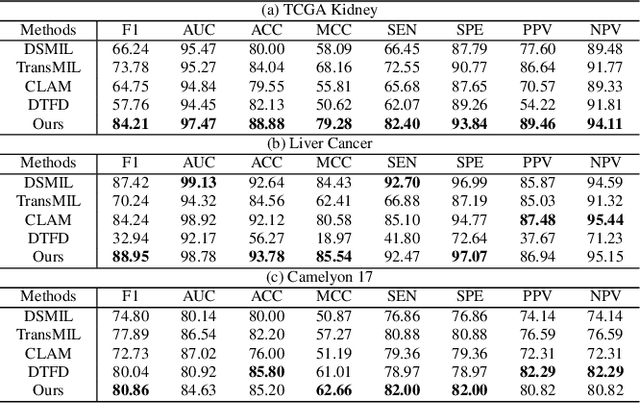
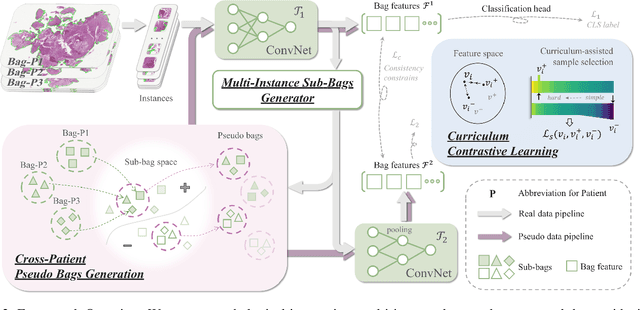
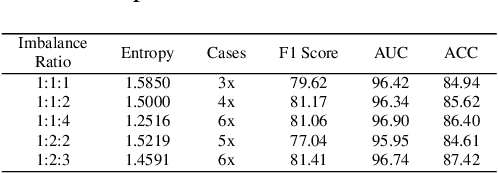
Abstract:Pathology computing has dramatically improved pathologists' workflow and diagnostic decision-making processes. Although computer-aided diagnostic systems have shown considerable value in whole slide image (WSI) analysis, the problem of multi-classification under sample imbalance remains an intractable challenge. To address this, we propose learning fine-grained information by generating sub-bags with feature distributions similar to the original WSIs. Additionally, we utilize a pseudo-bag generation algorithm to further leverage the abundant and redundant information in WSIs, allowing efficient training in unbalanced-sample multi-classification tasks. Furthermore, we introduce an affinity-based sample selection and curriculum contrastive learning strategy to enhance the stability of model representation learning. Unlike previous approaches, our framework transitions from learning bag-level representations to understanding and exploiting the feature distribution of multi-instance bags. Our method demonstrates significant performance improvements on three datasets, including tumor classification and lymph node metastasis. On average, it achieves a 4.39-point improvement in F1 score compared to the second-best method across the three tasks, underscoring its superior performance.
Multi-site, Multi-domain Airway Tree Modeling : A Public Benchmark for Pulmonary Airway Segmentation
Mar 10, 2023



Abstract:Open international challenges are becoming the de facto standard for assessing computer vision and image analysis algorithms. In recent years, new methods have extended the reach of pulmonary airway segmentation that is closer to the limit of image resolution. Since EXACT'09 pulmonary airway segmentation, limited effort has been directed to quantitative comparison of newly emerged algorithms driven by the maturity of deep learning based approaches and clinical drive for resolving finer details of distal airways for early intervention of pulmonary diseases. Thus far, public annotated datasets are extremely limited, hindering the development of data-driven methods and detailed performance evaluation of new algorithms. To provide a benchmark for the medical imaging community, we organized the Multi-site, Multi-domain Airway Tree Modeling (ATM'22), which was held as an official challenge event during the MICCAI 2022 conference. ATM'22 provides large-scale CT scans with detailed pulmonary airway annotation, including 500 CT scans (300 for training, 50 for validation, and 150 for testing). The dataset was collected from different sites and it further included a portion of noisy COVID-19 CTs with ground-glass opacity and consolidation. Twenty-three teams participated in the entire phase of the challenge and the algorithms for the top ten teams are reviewed in this paper. Quantitative and qualitative results revealed that deep learning models embedded with the topological continuity enhancement achieved superior performance in general. ATM'22 challenge holds as an open-call design, the training data and the gold standard evaluation are available upon successful registration via its homepage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge