Yogesh Verma
Positional Encoding meets Persistent Homology on Graphs
Jun 06, 2025Abstract:The local inductive bias of message-passing graph neural networks (GNNs) hampers their ability to exploit key structural information (e.g., connectivity and cycles). Positional encoding (PE) and Persistent Homology (PH) have emerged as two promising approaches to mitigate this issue. PE schemes endow GNNs with location-aware features, while PH methods enhance GNNs with multiresolution topological features. However, a rigorous theoretical characterization of the relative merits and shortcomings of PE and PH has remained elusive. We bridge this gap by establishing that neither paradigm is more expressive than the other, providing novel constructions where one approach fails but the other succeeds. Our insights inform the design of a novel learnable method, PiPE (Persistence-informed Positional Encoding), which is provably more expressive than both PH and PE. PiPE demonstrates strong performance across a variety of tasks (e.g., molecule property prediction, graph classification, and out-of-distribution generalization), thereby advancing the frontiers of graph representation learning. Code is available at https://github.com/Aalto-QuML/PIPE.
Robust Simulation-Based Inference under Missing Data via Neural Processes
Mar 03, 2025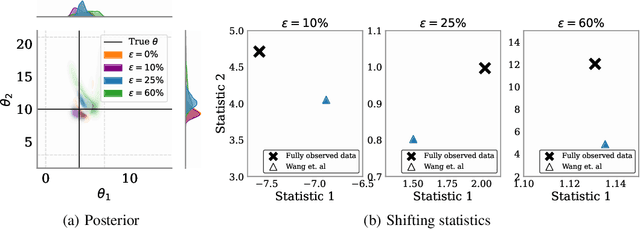

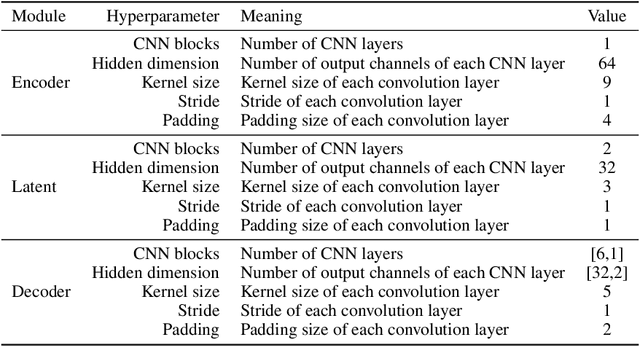
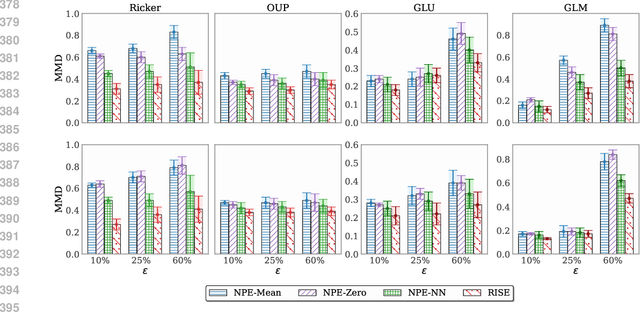
Abstract:Simulation-based inference (SBI) methods typically require fully observed data to infer parameters of models with intractable likelihood functions. However, datasets often contain missing values due to incomplete observations, data corruptions (common in astrophysics), or instrument limitations (e.g., in high-energy physics applications). In such scenarios, missing data must be imputed before applying any SBI method. We formalize the problem of missing data in SBI and demonstrate that naive imputation methods can introduce bias in the estimation of SBI posterior. We also introduce a novel amortized method that addresses this issue by jointly learning the imputation model and the inference network within a neural posterior estimation (NPE) framework. Extensive empirical results on SBI benchmarks show that our approach provides robust inference outcomes compared to standard baselines for varying levels of missing data. Moreover, we demonstrate the merits of our imputation model on two real-world bioactivity datasets (Adrenergic and Kinase assays). Code is available at https://github.com/Aalto-QuML/RISE.
Diffusion Twigs with Loop Guidance for Conditional Graph Generation
Oct 31, 2024



Abstract:We introduce a novel score-based diffusion framework named Twigs that incorporates multiple co-evolving flows for enriching conditional generation tasks. Specifically, a central or trunk diffusion process is associated with a primary variable (e.g., graph structure), and additional offshoot or stem processes are dedicated to dependent variables (e.g., graph properties or labels). A new strategy, which we call loop guidance, effectively orchestrates the flow of information between the trunk and the stem processes during sampling. This approach allows us to uncover intricate interactions and dependencies, and unlock new generative capabilities. We provide extensive experiments to demonstrate strong performance gains of the proposed method over contemporary baselines in the context of conditional graph generation, underscoring the potential of Twigs in challenging generative tasks such as inverse molecular design and molecular optimization.
Topological Neural Networks go Persistent, Equivariant, and Continuous
Jun 05, 2024



Abstract:Topological Neural Networks (TNNs) incorporate higher-order relational information beyond pairwise interactions, enabling richer representations than Graph Neural Networks (GNNs). Concurrently, topological descriptors based on persistent homology (PH) are being increasingly employed to augment the GNNs. We investigate the benefits of integrating these two paradigms. Specifically, we introduce TopNets as a broad framework that subsumes and unifies various methods in the intersection of GNNs/TNNs and PH such as (generalizations of) RePHINE and TOGL. TopNets can also be readily adapted to handle (symmetries in) geometric complexes, extending the scope of TNNs and PH to spatial settings. Theoretically, we show that PH descriptors can provably enhance the expressivity of simplicial message-passing networks. Empirically, (continuous and E(n)-equivariant extensions of) TopNets achieve strong performance across diverse tasks, including antibody design, molecular dynamics simulation, and drug property prediction.
ClimODE: Climate and Weather Forecasting with Physics-informed Neural ODEs
Apr 15, 2024Abstract:Climate and weather prediction traditionally relies on complex numerical simulations of atmospheric physics. Deep learning approaches, such as transformers, have recently challenged the simulation paradigm with complex network forecasts. However, they often act as data-driven black-box models that neglect the underlying physics and lack uncertainty quantification. We address these limitations with ClimODE, a spatiotemporal continuous-time process that implements a key principle of advection from statistical mechanics, namely, weather changes due to a spatial movement of quantities over time. ClimODE models precise weather evolution with value-conserving dynamics, learning global weather transport as a neural flow, which also enables estimating the uncertainty in predictions. Our approach outperforms existing data-driven methods in global and regional forecasting with an order of magnitude smaller parameterization, establishing a new state of the art.
Field-based Molecule Generation
Feb 24, 2024



Abstract:This work introduces FMG, a field-based model for drug-like molecule generation. We show how the flexibility of this method provides crucial advantages over the prevalent, point-cloud based methods, and achieves competitive molecular stability generation. We tackle optical isomerism (enantiomers), a previously omitted molecular property that is crucial for drug safety and effectiveness, and thus account for all molecular geometry aspects. We demonstrate how previous methods are invariant to a group of transformations that includes enantiomer pairs, leading them invariant to the molecular R and S configurations, while our field-based generative model captures this property.
AbODE: Ab Initio Antibody Design using Conjoined ODEs
May 31, 2023Abstract:Antibodies are Y-shaped proteins that neutralize pathogens and constitute the core of our adaptive immune system. De novo generation of new antibodies that target specific antigens holds the key to accelerating vaccine discovery. However, this co-design of the amino acid sequence and the 3D structure subsumes and accentuates some central challenges from multiple tasks, including protein folding (sequence to structure), inverse folding (structure to sequence), and docking (binding). We strive to surmount these challenges with a new generative model AbODE that extends graph PDEs to accommodate both contextual information and external interactions. Unlike existing approaches, AbODE uses a single round of full-shot decoding and elicits continuous differential attention that encapsulates and evolves with latent interactions within the antibody as well as those involving the antigen. We unravel fundamental connections between AbODE and temporal networks as well as graph-matching networks. The proposed model significantly outperforms existing methods on standard metrics across benchmarks.
Modular Flows: Differential Molecular Generation
Oct 13, 2022
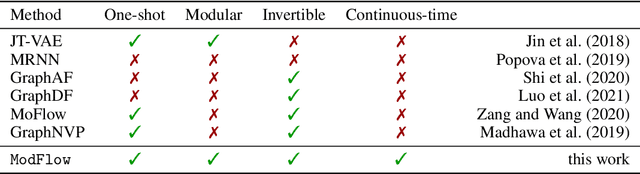


Abstract:Generating new molecules is fundamental to advancing critical applications such as drug discovery and material synthesis. Flows can generate molecules effectively by inverting the encoding process, however, existing flow models either require artifactual dequantization or specific node/edge orderings, lack desiderata such as permutation invariance, or induce discrepancy between the encoding and the decoding steps that necessitates post hoc validity correction. We circumvent these issues with novel continuous normalizing E(3)-equivariant flows, based on a system of node ODEs coupled as a graph PDE, that repeatedly reconcile locally toward globally aligned densities. Our models can be cast as message-passing temporal networks, and result in superlative performance on the tasks of density estimation and molecular generation. In particular, our generated samples achieve state-of-the-art on both the standard QM9 and ZINC250K benchmarks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge