Yoeri Poels
Plasma State Monitoring and Disruption Characterization using Multimodal VAEs
Apr 24, 2025Abstract:When a plasma disrupts in a tokamak, significant heat and electromagnetic loads are deposited onto the surrounding device components. These forces scale with plasma current and magnetic field strength, making disruptions one of the key challenges for future devices. Unfortunately, disruptions are not fully understood, with many different underlying causes that are difficult to anticipate. Data-driven models have shown success in predicting them, but they only provide limited interpretability. On the other hand, large-scale statistical analyses have been a great asset to understanding disruptive patterns. In this paper, we leverage data-driven methods to find an interpretable representation of the plasma state for disruption characterization. Specifically, we use a latent variable model to represent diagnostic measurements as a low-dimensional, latent representation. We build upon the Variational Autoencoder (VAE) framework, and extend it for (1) continuous projections of plasma trajectories; (2) a multimodal structure to separate operating regimes; and (3) separation with respect to disruptive regimes. Subsequently, we can identify continuous indicators for the disruption rate and the disruptivity based on statistical properties of measurement data. The proposed method is demonstrated using a dataset of approximately 1600 TCV discharges, selecting for flat-top disruptions or regular terminations. We evaluate the method with respect to (1) the identified disruption risk and its correlation with other plasma properties; (2) the ability to distinguish different types of disruptions; and (3) downstream analyses. For the latter, we conduct a demonstrative study on identifying parameters connected to disruptions using counterfactual-like analysis. Overall, the method can adequately identify distinct operating regimes characterized by varying proximity to disruptions in an interpretable manner.
Robust Confinement State Classification with Uncertainty Quantification through Ensembled Data-Driven Methods
Feb 24, 2025Abstract:Maximizing fusion performance in tokamaks relies on high energy confinement, often achieved through distinct operating regimes. The automated labeling of these confinement states is crucial to enable large-scale analyses or for real-time control applications. While this task becomes difficult to automate near state transitions or in marginal scenarios, much success has been achieved with data-driven models. However, these methods generally provide predictions as point estimates, and cannot adequately deal with missing and/or broken input signals. To enable wide-range applicability, we develop methods for confinement state classification with uncertainty quantification and model robustness. We focus on off-line analysis for TCV discharges, distinguishing L-mode, H-mode, and an in-between dithering phase (D). We propose ensembling data-driven methods on two axes: model formulations and feature sets. The former considers a dynamic formulation based on a recurrent Fourier Neural Operator-architecture and a static formulation based on gradient-boosted decision trees. These models are trained using multiple feature groupings categorized by diagnostic system or physical quantity. A dataset of 302 TCV discharges is fully labeled, and will be publicly released. We evaluate our method quantitatively using Cohen's kappa coefficient for predictive performance and the Expected Calibration Error for the uncertainty calibration. Furthermore, we discuss performance using a variety of common and alternative scenarios, the performance of individual components, out-of-distribution performance, cases of broken or missing signals, and evaluate conditionally-averaged behavior around different state transitions. Overall, the proposed method can distinguish L, D and H-mode with high performance, can cope with missing or broken signals, and provides meaningful uncertainty estimates.
Learning Plasma Dynamics and Robust Rampdown Trajectories with Predict-First Experiments at TCV
Feb 17, 2025Abstract:The rampdown in tokamak operations is a difficult to simulate phase during which the plasma is often pushed towards multiple instability limits. To address this challenge, and reduce the risk of disrupting operations, we leverage recent advances in Scientific Machine Learning (SciML) to develop a neural state-space model (NSSM) that predicts plasma dynamics during Tokamak \`a Configuration Variable (TCV) rampdowns. By integrating simple physics structure and data-driven models, the NSSM efficiently learns plasma dynamics during the rampdown from a modest dataset of 311 pulses with only five pulses in the reactor relevant high performance regime. The NSSM is parallelized across uncertainties, and reinforcement learning (RL) is applied to design trajectories that avoid multiple instability limits with high probability. Experiments at TCV ramping down high performance plasmas show statistically significant improvements in current and energy at plasma termination, with improvements in speed through continuous re-training. A predict-first experiment, increasing plasma current by 20\% from baseline, demonstrates the NSSM's ability to make small extrapolations with sufficient accuracy to design trajectories that successfully terminate the pulse. The developed approach paves the way for designing tokamak controls with robustness to considerable uncertainty, and demonstrates the relevance of the SciML approach to learning plasma dynamics for rapidly developing robust trajectories and controls during the incremental campaigns of upcoming burning plasma tokamaks.
Accelerating Simulation of Two-Phase Flows with Neural PDE Surrogates
May 27, 2024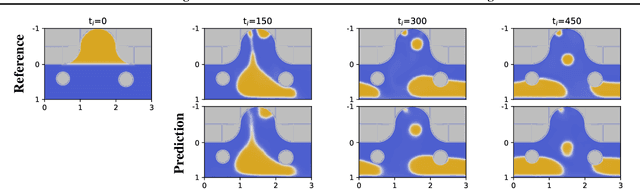
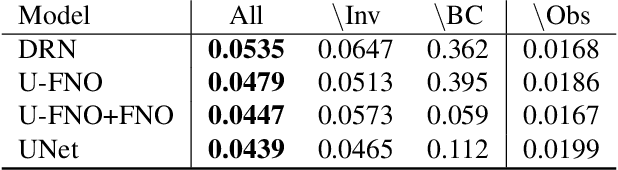
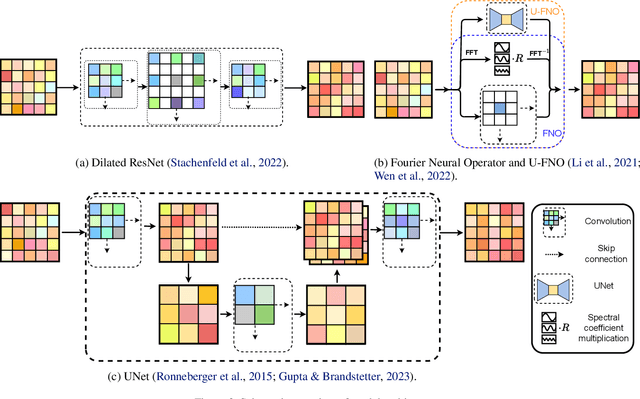
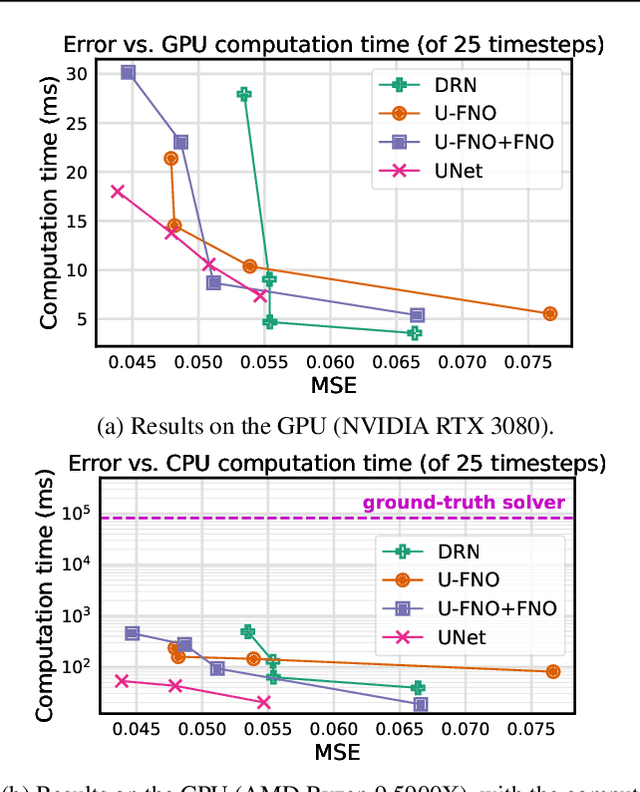
Abstract:Simulation is a powerful tool to better understand physical systems, but generally requires computationally expensive numerical methods. Downstream applications of such simulations can become computationally infeasible if they require many forward solves, for example in the case of inverse design with many degrees of freedom. In this work, we investigate and extend neural PDE solvers as a tool to aid in scaling simulations for two-phase flow problems, and simulations of oil expulsion from a pore specifically. We extend existing numerical methods for this problem to a more complex setting involving varying geometries of the domain to generate a challenging dataset. Further, we investigate three prominent neural PDE solver methods, namely the UNet, DRN and U-FNO, and extend them for characteristics of the oil-expulsion problem: (1) spatial conditioning on the geometry; (2) periodicity in the boundary; (3) approximate mass conservation. We scale all methods and benchmark their speed-accuracy trade-off, evaluate qualitative properties, and perform an ablation study. We find that the investigated methods can accurately model the droplet dynamics with up to three orders of magnitude speed-up, that our extensions improve performance over the baselines, and that the introduced varying geometries constitute a significantly more challenging setting over the previously considered oil expulsion problem.
Fast Dynamic 1D Simulation of Divertor Plasmas with Neural PDE Surrogates
May 30, 2023Abstract:Managing divertor plasmas is crucial for operating reactor scale tokamak devices due to heat and particle flux constraints on the divertor target. Simulation is an important tool to understand and control these plasmas, however, for real-time applications or exhaustive parameter scans only simple approximations are currently fast enough. We address this lack of fast simulators using neural PDE surrogates, data-driven neural network-based surrogate models trained using solutions generated with a classical numerical method. The surrogate approximates a time-stepping operator that evolves the full spatial solution of a reference physics-based model over time. We use DIV1D, a 1D dynamic model of the divertor plasma, as reference model to generate data. DIV1D's domain covers a 1D heat flux tube from the X-point (upstream) to the target. We simulate a realistic TCV divertor plasma with dynamics induced by upstream density ramps and provide an exploratory outlook towards fast transients. State-of-the-art neural PDE surrogates are evaluated in a common framework and extended for properties of the DIV1D data. We evaluate (1) the speed-accuracy trade-off; (2) recreating non-linear behavior; (3) data efficiency; and (4) parameter inter- and extrapolation. Once trained, neural PDE surrogates can faithfully approximate DIV1D's divertor plasma dynamics at sub real-time computation speeds: In the proposed configuration, 2ms of plasma dynamics can be computed in $\approx$0.63ms of wall-clock time, several orders of magnitude faster than DIV1D.
Equivariant Neural Simulators for Stochastic Spatiotemporal Dynamics
May 23, 2023



Abstract:Neural networks are emerging as a tool for scalable data-driven simulation of high-dimensional dynamical systems, especially in settings where numerical methods are infeasible or computationally expensive. Notably, it has been shown that incorporating domain symmetries in deterministic neural simulators can substantially improve their accuracy, sample efficiency, and parameter efficiency. However, to incorporate symmetries in probabilistic neural simulators that can simulate stochastic phenomena, we need a model that produces equivariant distributions over trajectories, rather than equivariant function approximations. In this paper, we propose Equivariant Probabilistic Neural Simulation (EPNS), a framework for autoregressive probabilistic modeling of equivariant distributions over system evolutions. We use EPNS to design models for a stochastic n-body system and stochastic cellular dynamics. Our results show that EPNS considerably outperforms existing neural network-based methods for probabilistic simulation. More specifically, we demonstrate that incorporating equivariance in EPNS improves simulation quality, data efficiency, rollout stability, and uncertainty quantification. We conclude that EPNS is a promising method for efficient and effective data-driven probabilistic simulation in a diverse range of domains.
Towards Learned Simulators for Cell Migration
Oct 02, 2022



Abstract:Simulators driven by deep learning are gaining popularity as a tool for efficiently emulating accurate but expensive numerical simulators. Successful applications of such neural simulators can be found in the domains of physics, chemistry, and structural biology, amongst others. Likewise, a neural simulator for cellular dynamics can augment lab experiments and traditional computational methods to enhance our understanding of a cell's interaction with its physical environment. In this work, we propose an autoregressive probabilistic model that can reproduce spatiotemporal dynamics of single cell migration, traditionally simulated with the Cellular Potts model. We observe that standard single-step training methods do not only lead to inconsistent rollout stability, but also fail to accurately capture the stochastic aspects of the dynamics, and we propose training strategies to mitigate these issues. Our evaluation on two proof-of-concept experimental scenarios shows that neural methods have the potential to faithfully simulate stochastic cellular dynamics at least an order of magnitude faster than a state-of-the-art implementation of the Cellular Potts model.
VAE-CE: Visual Contrastive Explanation using Disentangled VAEs
Aug 20, 2021



Abstract:The goal of a classification model is to assign the correct labels to data. In most cases, this data is not fully described by the given set of labels. Often a rich set of meaningful concepts exist in the domain that can much more precisely describe each datapoint. Such concepts can also be highly useful for interpreting the model's classifications. In this paper we propose a model, denoted as Variational Autoencoder-based Contrastive Explanation (VAE-CE), that represents data with high-level concepts and uses this representation for both classification and generating explanations. The explanations are produced in a contrastive manner, conveying why a datapoint is assigned to one class rather than an alternative class. An explanation is specified as a set of transformations of the input datapoint, with each step depicting a concept changing towards the contrastive class. We build the model using a disentangled VAE, extended with a new supervised method for disentangling individual dimensions. An analysis on synthetic data and MNIST shows that the approaches to both disentanglement and explanation provide benefits over other methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge