Xiwei Liu
Towards Robust Visual Continual Learning with Multi-Prototype Supervision
Sep 19, 2025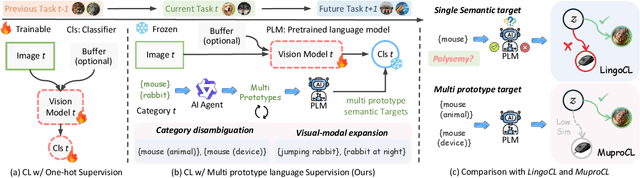
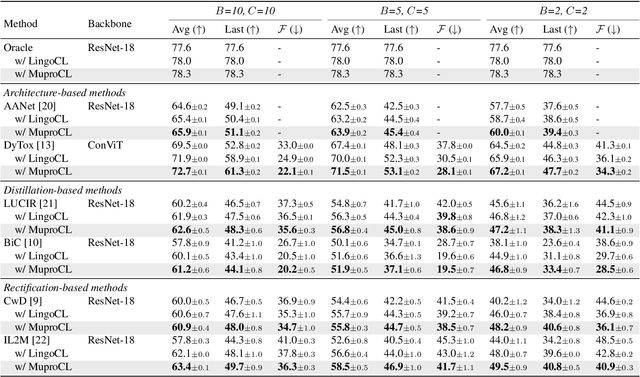
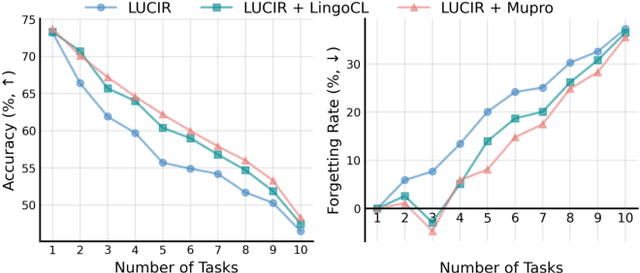
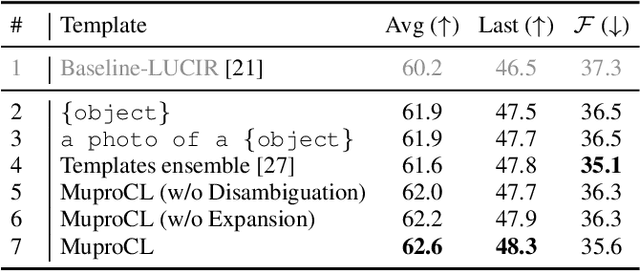
Abstract:Language-guided supervision, which utilizes a frozen semantic target from a Pretrained Language Model (PLM), has emerged as a promising paradigm for visual Continual Learning (CL). However, relying on a single target introduces two critical limitations: 1) semantic ambiguity, where a polysemous category name results in conflicting visual representations, and 2) intra-class visual diversity, where a single prototype fails to capture the rich variety of visual appearances within a class. To this end, we propose MuproCL, a novel framework that replaces the single target with multiple, context-aware prototypes. Specifically, we employ a lightweight LLM agent to perform category disambiguation and visual-modal expansion to generate a robust set of semantic prototypes. A LogSumExp aggregation mechanism allows the vision model to adaptively align with the most relevant prototype for a given image. Extensive experiments across various CL baselines demonstrate that MuproCL consistently enhances performance and robustness, establishing a more effective path for language-guided continual learning.
SSS: Semi-Supervised SAM-2 with Efficient Prompting for Medical Imaging Segmentation
Jun 10, 2025Abstract:In the era of information explosion, efficiently leveraging large-scale unlabeled data while minimizing the reliance on high-quality pixel-level annotations remains a critical challenge in the field of medical imaging. Semi-supervised learning (SSL) enhances the utilization of unlabeled data by facilitating knowledge transfer, significantly improving the performance of fully supervised models and emerging as a highly promising research direction in medical image analysis. Inspired by the ability of Vision Foundation Models (e.g., SAM-2) to provide rich prior knowledge, we propose SSS (Semi-Supervised SAM-2), a novel approach that leverages SAM-2's robust feature extraction capabilities to uncover latent knowledge in unlabeled medical images, thus effectively enhancing feature support for fully supervised medical image segmentation. Specifically, building upon the single-stream "weak-to-strong" consistency regularization framework, this paper introduces a Discriminative Feature Enhancement (DFE) mechanism to further explore the feature discrepancies introduced by various data augmentation strategies across multiple views. By leveraging feature similarity and dissimilarity across multi-scale augmentation techniques, the method reconstructs and models the features, thereby effectively optimizing the salient regions. Furthermore, a prompt generator is developed that integrates Physical Constraints with a Sliding Window (PCSW) mechanism to generate input prompts for unlabeled data, fulfilling SAM-2's requirement for additional prompts. Extensive experiments demonstrate the superiority of the proposed method for semi-supervised medical image segmentation on two multi-label datasets, i.e., ACDC and BHSD. Notably, SSS achieves an average Dice score of 53.15 on BHSD, surpassing the previous state-of-the-art method by +3.65 Dice. Code will be available at https://github.com/AIGeeksGroup/SSS.
J-Invariant Volume Shuffle for Self-Supervised Cryo-Electron Tomogram Denoising on Single Noisy Volume
Nov 22, 2024



Abstract:Cryo-Electron Tomography (Cryo-ET) enables detailed 3D visualization of cellular structures in near-native states but suffers from low signal-to-noise ratio due to imaging constraints. Traditional denoising methods and supervised learning approaches often struggle with complex noise patterns and the lack of paired datasets. Self-supervised methods, which utilize noisy input itself as a target, have been studied; however, existing Cryo-ET self-supervised denoising methods face significant challenges due to losing information during training and the learned incomplete noise patterns. In this paper, we propose a novel self-supervised learning model that denoises Cryo-ET volumetric images using a single noisy volume. Our method features a U-shape J-invariant blind spot network with sparse centrally masked convolutions, dilated channel attention blocks, and volume unshuffle/shuffle technique. The volume-unshuffle/shuffle technique expands receptive fields and utilizes multi-scale representations, significantly improving noise reduction and structural preservation. Experimental results demonstrate that our approach achieves superior performance compared to existing methods, advancing Cryo-ET data processing for structural biology research
FIAS: Feature Imbalance-Aware Medical Image Segmentation with Dynamic Fusion and Mixing Attention
Nov 16, 2024
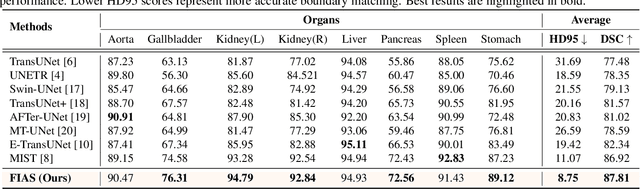

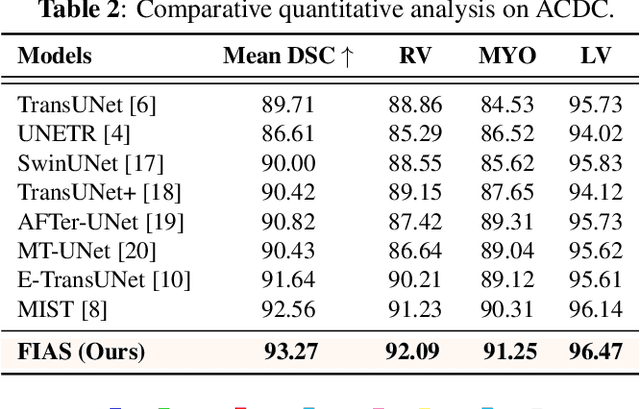
Abstract:With the growing application of transformer in computer vision, hybrid architecture that combine convolutional neural networks (CNNs) and transformers demonstrates competitive ability in medical image segmentation. However, direct fusion of features from CNNs and transformers often leads to feature imbalance and redundant information. To address these issues, we propose a Feaure Imbalance-Aware Segmentation (FIAS) network, which incorporates a dual-path encoder and a novel Mixing Attention (MixAtt) decoder. The dual-branches encoder integrates a DilateFormer for long-range global feature extraction and a Depthwise Multi-Kernel (DMK) convolution for capturing fine-grained local details. A Context-Aware Fusion (CAF) block dynamically balances the contribution of these global and local features, preventing feature imbalance. The MixAtt decoder further enhances segmentation accuracy by combining self-attention and Monte Carlo attention, enabling the model to capture both small details and large-scale dependencies. Experimental results on the Synapse multi-organ and ACDC datasets demonstrate the strong competitiveness of our approach in medical image segmentation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge