Xiangyu Xiong
Distance Guided Generative Adversarial Network for Explainable Binary Classifications
Dec 29, 2023Abstract:Despite the potential benefits of data augmentation for mitigating the data insufficiency, traditional augmentation methods primarily rely on the prior intra-domain knowledge. On the other hand, advanced generative adversarial networks (GANs) generate inter-domain samples with limited variety. These previous methods make limited contributions to describing the decision boundaries for binary classification. In this paper, we propose a distance guided GAN (DisGAN) which controls the variation degrees of generated samples in the hyperplane space. Specifically, we instantiate the idea of DisGAN by combining two ways. The first way is vertical distance GAN (VerDisGAN) where the inter-domain generation is conditioned on the vertical distances. The second way is horizontal distance GAN (HorDisGAN) where the intra-domain generation is conditioned on the horizontal distances. Furthermore, VerDisGAN can produce the class-specific regions by mapping the source images to the hyperplane. Experimental results show that DisGAN consistently outperforms the GAN-based augmentation methods with explainable binary classification. The proposed method can apply to different classification architectures and has potential to extend to multi-class classification.
A Parameterized Generative Adversarial Network Using Cyclic Projection for Explainable Medical Image Classification
Dec 07, 2023
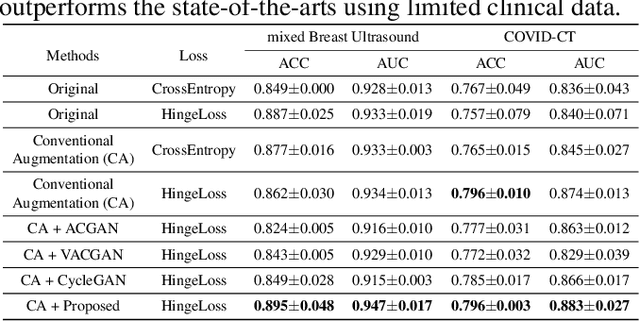

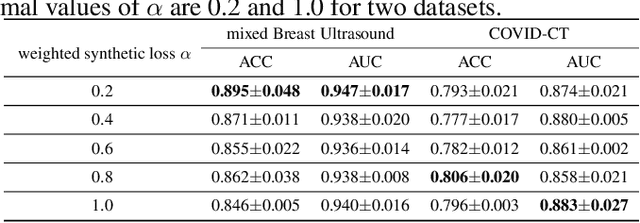
Abstract:Although current data augmentation methods are successful to alleviate the data insufficiency, conventional augmentation are primarily intra-domain while advanced generative adversarial networks (GANs) generate images remaining uncertain, particularly in small-scale datasets. In this paper, we propose a parameterized GAN (ParaGAN) that effectively controls the changes of synthetic samples among domains and highlights the attention regions for downstream classification. Specifically, ParaGAN incorporates projection distance parameters in cyclic projection and projects the source images to the decision boundary to obtain the class-difference maps. Our experiments show that ParaGAN can consistently outperform the existing augmentation methods with explainable classification on two small-scale medical datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge