William S. York
Identification of 1H-NMR Spectra of Xyloglucan Oligosaccharides: A Comparative Study of Artificial Neural Networks and Bayesian Classification Using Nonparametric Density Estimation
Jul 30, 2020Abstract:Proton nuclear magnetic resonance (1H-NMR) is a widely used tool for chemical structural analysis. However, 1H-NMR spectra suffer from natural aberrations that render computer-assisted automated identification of these spectra difficult, and at times impossible. Previous efforts have successfully implemented instrument dependent or conditional identification of these spectra. In this paper, we report the first instrument independent computer-assisted automated identification system for a group of complex carbohydrates known as the xyloglucan oligosaccharides. The developed system is also implemented on the world wide web (http://www.ccrc.uga.edu) as part of an identification package called the CCRC-Net and is intended to recognize any submitted 1H-NMR spectrum of these structures with reasonable signal-to-noise ratio, recorded on any 500 MHz NMR instrument. The system uses Artificial Neural Networks (ANNs) technology and is insensitive to the instrument and environment-dependent variations in 1H-NMR spectroscopy. In this paper, comparative results of the ANN engine versus a multidimensional Bayes' classifier is also presented.
* 6 pages. Published in IEEE ICAI99
GELATO and SAGE: An Integrated Framework for MS Annotation
Jan 08, 2016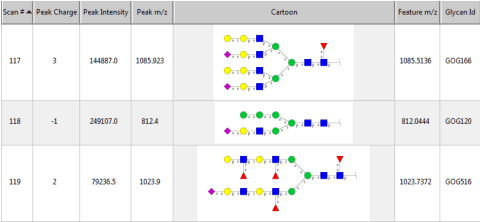
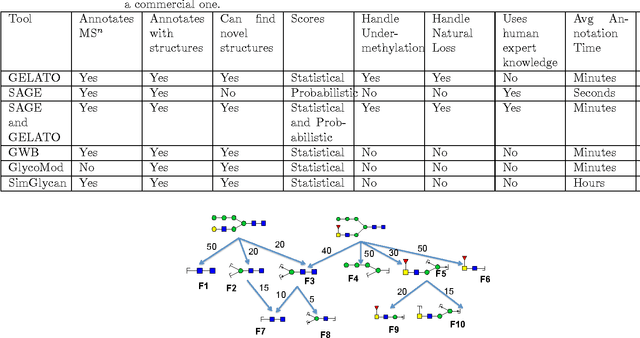
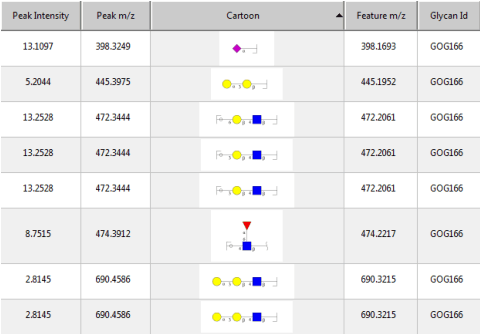
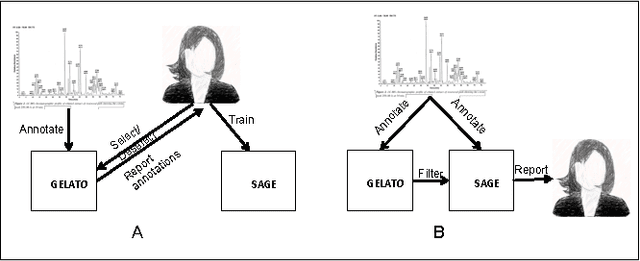
Abstract:Several algorithms and tools have been developed to (semi) automate the process of glycan identification by interpreting Mass Spectrometric data. However, each has limitations when annotating MSn data with thousands of MS spectra using uncurated public databases. Moreover, the existing tools are not designed to manage MSn data where n > 2. We propose a novel software package to automate the annotation of tandem MS data. This software consists of two major components. The first, is a free, semi-automated MSn data interpreter called the Glycomic Elucidation and Annotation Tool (GELATO). This tool extends and automates the functionality of existing open source projects, namely, GlycoWorkbench (GWB) and GlycomeDB. The second is a machine learning model called Smart Anotation Enhancement Graph (SAGE), which learns the behavior of glycoanalysts to select annotations generated by GELATO that emulate human interpretation of the spectra.
Mining Massive Hierarchical Data Using a Scalable Probabilistic Graphical Model
Dec 28, 2015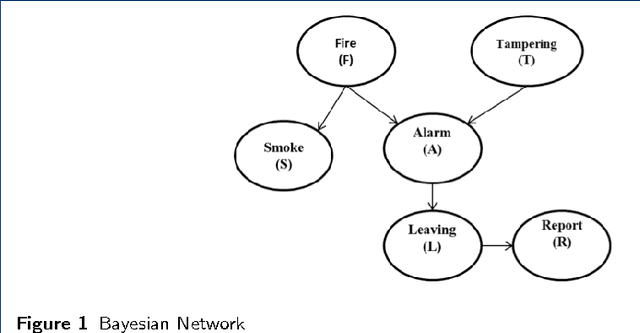
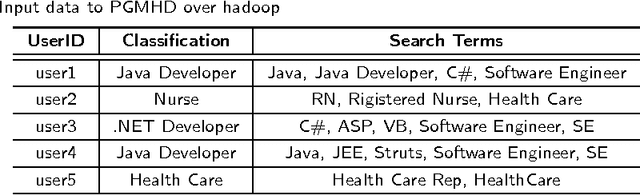
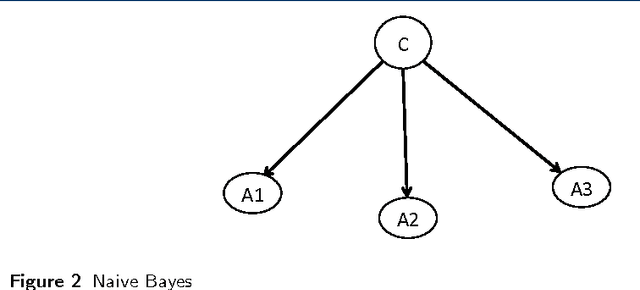

Abstract:Probabilistic Graphical Models (PGM) are very useful in the fields of machine learning and data mining. The crucial limitation of those models,however, is the scalability. The Bayesian Network, which is one of the most common PGMs used in machine learning and data mining, demonstrates this limitation when the training data consists of random variables, each of them has a large set of possible values. In the big data era, one would expect new extensions to the existing PGMs to handle the massive amount of data produced these days by computers, sensors and other electronic devices. With hierarchical data - data that is arranged in a treelike structure with several levels - one would expect to see hundreds of thousands or millions of values distributed over even just a small number of levels. When modeling this kind of hierarchical data across large data sets, Bayesian Networks become infeasible for representing the probability distributions. In this paper we introduce an extension to Bayesian Networks to handle massive sets of hierarchical data in a reasonable amount of time and space. The proposed model achieves perfect precision of 1.0 and high recall of 0.93 when it is used as multi-label classifier for the annotation of mass spectrometry data. On another data set of 1.5 billion search logs provided by CareerBuilder.com the model was able to predict latent semantic relationships between search keywords with accuracy up to 0.80.
PGMHD: A Scalable Probabilistic Graphical Model for Massive Hierarchical Data Problems
Aug 19, 2014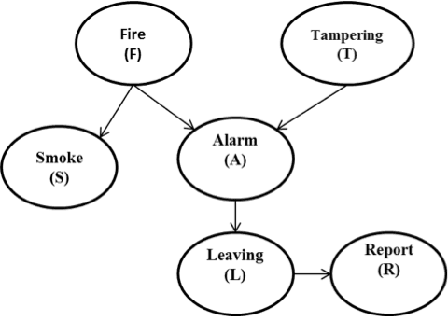
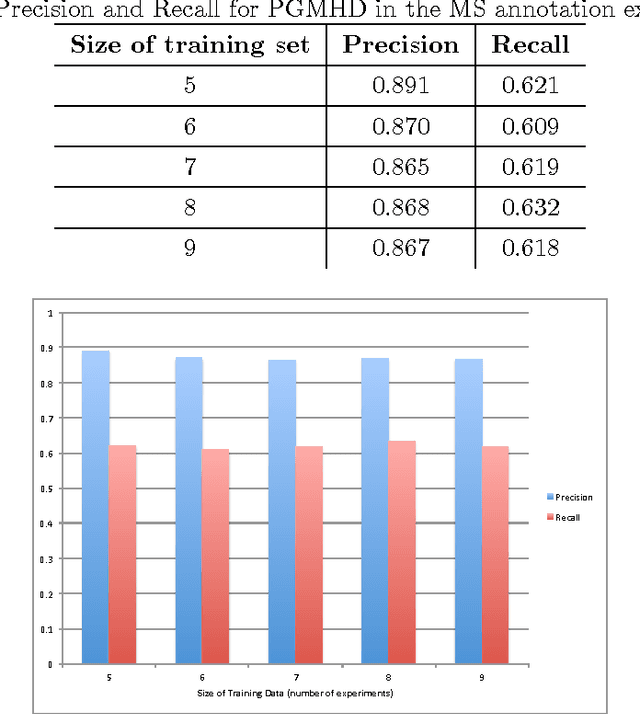
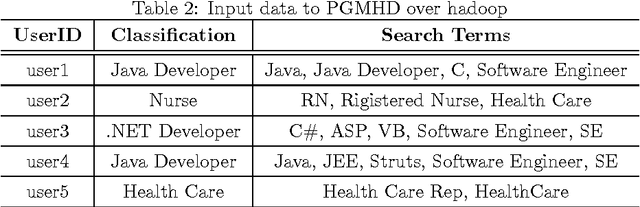
Abstract:In the big data era, scalability has become a crucial requirement for any useful computational model. Probabilistic graphical models are very useful for mining and discovering data insights, but they are not scalable enough to be suitable for big data problems. Bayesian Networks particularly demonstrate this limitation when their data is represented using few random variables while each random variable has a massive set of values. With hierarchical data - data that is arranged in a treelike structure with several levels - one would expect to see hundreds of thousands or millions of values distributed over even just a small number of levels. When modeling this kind of hierarchical data across large data sets, Bayesian networks become infeasible for representing the probability distributions for the following reasons: i) Each level represents a single random variable with hundreds of thousands of values, ii) The number of levels is usually small, so there are also few random variables, and iii) The structure of the network is predefined since the dependency is modeled top-down from each parent to each of its child nodes, so the network would contain a single linear path for the random variables from each parent to each child node. In this paper we present a scalable probabilistic graphical model to overcome these limitations for massive hierarchical data. We believe the proposed model will lead to an easily-scalable, more readable, and expressive implementation for problems that require probabilistic-based solutions for massive amounts of hierarchical data. We successfully applied this model to solve two different challenging probabilistic-based problems on massive hierarchical data sets for different domains, namely, bioinformatics and latent semantic discovery over search logs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge