Homayoun Valafar
Computer Science and Engineering Department, University of South Carolina
Automated Measurement of Vascular Calcification in Femoral Endarterectomy Patients Using Deep Learning
Nov 27, 2023Abstract:Atherosclerosis, a chronic inflammatory disease affecting the large arteries, presents a global health risk. Accurate analysis of diagnostic images, like computed tomographic angiograms (CTAs), is essential for staging and monitoring the progression of atherosclerosis-related conditions, including peripheral arterial disease (PAD). However, manual analysis of CTA images is time-consuming and tedious. To address this limitation, we employed a deep learning model to segment the vascular system in CTA images of PAD patients undergoing femoral endarterectomy surgery and to measure vascular calcification from the left renal artery to the patella. Utilizing proprietary CTA images of 27 patients undergoing femoral endarterectomy surgery provided by Prisma Health Midlands, we developed a Deep Neural Network (DNN) model to first segment the arterial system, starting from the descending aorta to the patella, and second, to provide a metric of arterial calcification. Our designed DNN achieved 83.4% average Dice accuracy in segmenting arteries from aorta to patella, advancing the state-of-the-art by 0.8%. Furthermore, our work is the first to present a robust statistical analysis of automated calcification measurement in the lower extremities using deep learning, attaining a Mean Absolute Percentage Error (MAPE) of 9.5% and a correlation coefficient of 0.978 between automated and manual calcification scores. These findings underscore the potential of deep learning techniques as a rapid and accurate tool for medical professionals to assess calcification in the abdominal aorta and its branches above the patella. The developed DNN model and related documentation in this project are available at GitHub page at https://github.com/pip-alireza/DeepCalcScoring.
* Published in MDPI Diagnostic journal, the code can be accessed via the GitHub link in the paper
TransONet: Automatic Segmentation of Vasculature in Computed Tomographic Angiograms Using Deep Learning
Nov 17, 2023Abstract:Pathological alterations in the human vascular system underlie many chronic diseases, such as atherosclerosis and aneurysms. However, manually analyzing diagnostic images of the vascular system, such as computed tomographic angiograms (CTAs) is a time-consuming and tedious process. To address this issue, we propose a deep learning model to segment the vascular system in CTA images of patients undergoing surgery for peripheral arterial disease (PAD). Our study focused on accurately segmenting the vascular system (1) from the descending thoracic aorta to the iliac bifurcation and (2) from the descending thoracic aorta to the knees in CTA images using deep learning techniques. Our approach achieved average Dice accuracies of 93.5% and 80.64% in test dataset for (1) and (2), respectively, highlighting its high accuracy and potential clinical utility. These findings demonstrate the use of deep learning techniques as a valuable tool for medical professionals to analyze the health of the vascular system efficiently and accurately. Please visit the GitHub page for this paper at https://github.com/pip-alireza/TransOnet.
nD-PDPA: nDimensional Probability Density Profile Analysis
Apr 05, 2023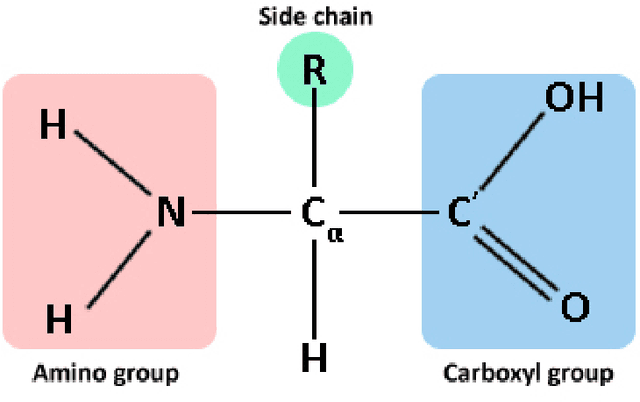
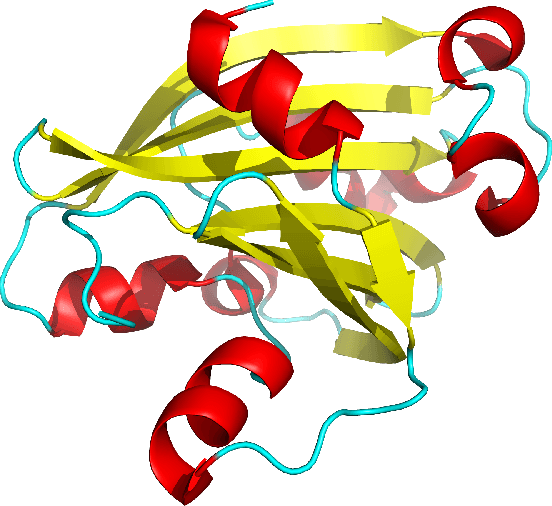
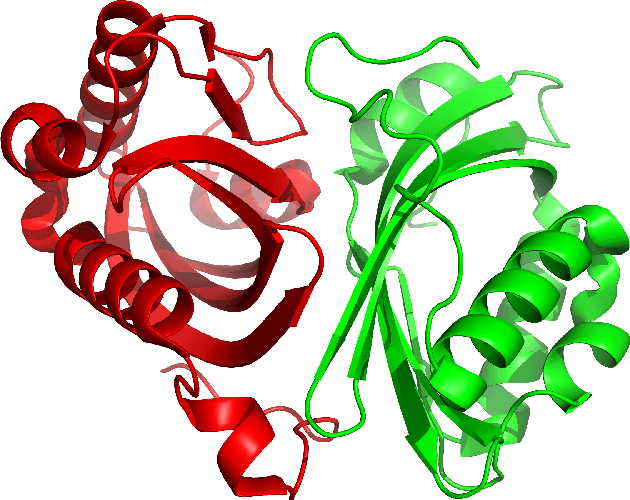
Abstract:Despite the recent advances in various Structural Genomics Projects, a large gap remains between the number of sequenced and structurally characterized proteins. Some reasons for this discrepancy include technical difficulties, labor, and the cost related to determining a structure by experimental methods such as NMR spectroscopy. Several computational methods have been developed to expand the applicability of NMR spectroscopy by addressing temporal and economical problems more efficiently. While these methods demonstrate successful outcomes to solve more challenging and structurally novel proteins, the cost has not been reduced significantly. Probability Density Profile Analysis (PDPA) has been previously introduced by our lab to directly address the economics of structure determination of routine proteins and the identification of novel structures from a minimal set of unassigned NMR data. 2D-PDPA (in which 2D denotes incorporation of data from two alignment media) has been successful in identifying the structural homolog of an unknown protein within a library of ~1000 decoy structures. In order to further expand the selectivity and sensitivity of PDPA, the incorporation of additional data was necessary. However, the expansion of the original PDPA approach was limited by its computational requirements where the inclusion of additional data would render it computationally intractable. Here we present the most recent developments of PDPA method (nD-PDPA: n Dimensional Probability Density Profile Analysis) that eliminate 2D-PDPA's computational limitations, and allows inclusion of RDC data from multiple vector types in multiple alignment media.
Application of Dimensional Reduction in Artificial Neural Networks to Improve Emergency Department Triage During Chemical Mass Casualty Incidents
Apr 01, 2022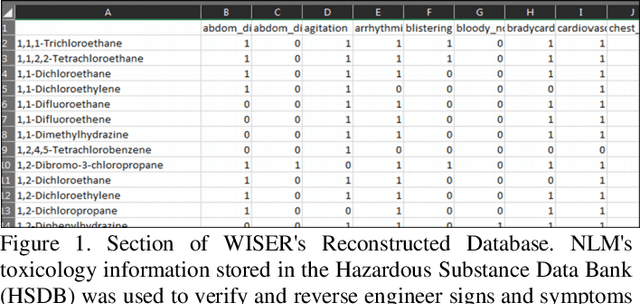
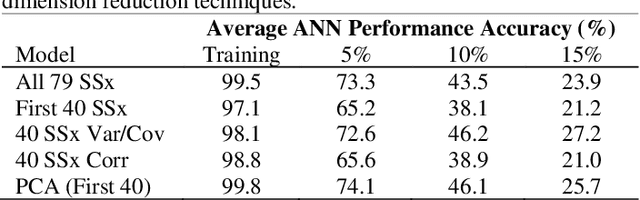
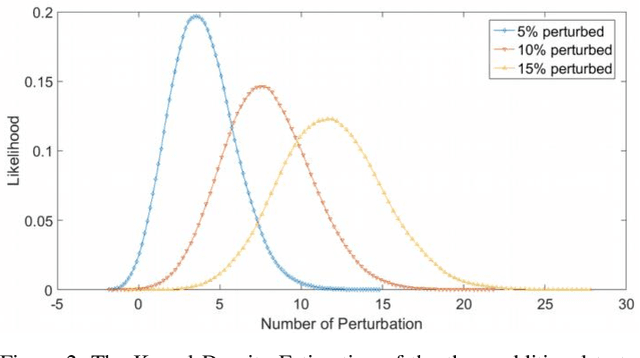
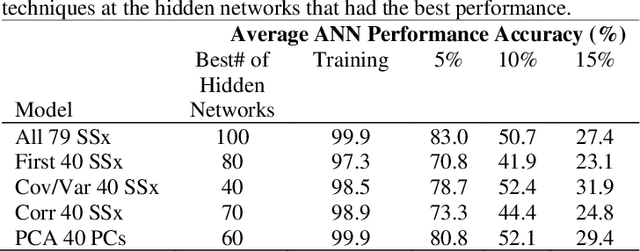
Abstract:Chemical Mass Casualty Incidents (MCI) place a heavy burden on hospital staff and resources. Machine Learning (ML) tools can provide efficient decision support to caregivers. However, ML models require large volumes of data for the most accurate results, which is typically not feasible in the chaotic nature of a chemical MCI. This study examines the application of four statistical dimension reduction techniques: Random Selection, Covariance/Variance, Pearson's Linear Correlation, and Principle Component Analysis to reduce a dataset of 311 hazardous chemicals and 79 related signs and symptoms (SSx). An Artificial Neural Network pipeline was developed to create comparative models. Results show that the number of signs and symptoms needed to determine a chemical culprit can be reduced to nearly 40 SSx without losing significant model accuracy. Evidence also suggests that the application of dimension reduction methods can improve ANN model performance accuracy.
Application of Machine Learning to Sleep Stage Classification
Nov 04, 2021
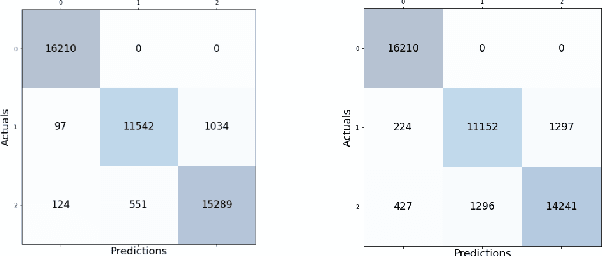
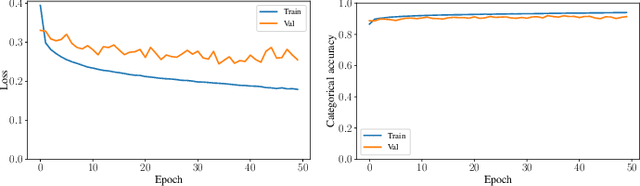
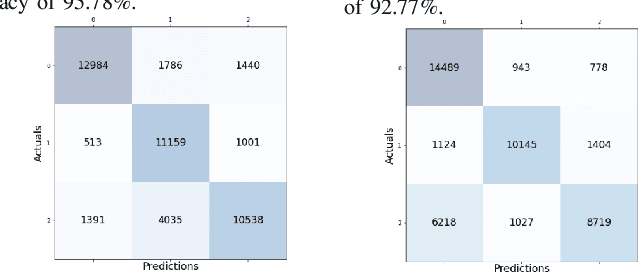
Abstract:Sleep studies are imperative to recapitulate phenotypes associated with sleep loss and uncover mechanisms contributing to psychopathology. Most often, investigators manually classify the polysomnography into vigilance states, which is time-consuming, requires extensive training, and is prone to inter-scorer variability. While many works have successfully developed automated vigilance state classifiers based on multiple EEG channels, we aim to produce an automated and open-access classifier that can reliably predict vigilance state based on a single cortical electroencephalogram (EEG) from rodents to minimize the disadvantages that accompany tethering small animals via wires to computer programs. Approximately 427 hours of continuously monitored EEG, electromyogram (EMG), and activity were labeled by a domain expert out of 571 hours of total data. Here we evaluate the performance of various machine learning techniques on classifying 10-second epochs into one of three discrete classes: paradoxical, slow-wave, or wake. Our investigations include Decision Trees, Random Forests, Naive Bayes Classifiers, Logistic Regression Classifiers, and Artificial Neural Networks. These methodologies have achieved accuracies ranging from approximately 74% to approximately 96%. Most notably, the Random Forest and the ANN achieved remarkable accuracies of 95.78% and 93.31%, respectively. Here we have shown the potential of various machine learning classifiers to automatically, accurately, and reliably classify vigilance states based on a single EEG reading and a single EMG reading.
Application of Machine Learning in Early Recommendation of Cardiac Resynchronization Therapy
Sep 13, 2021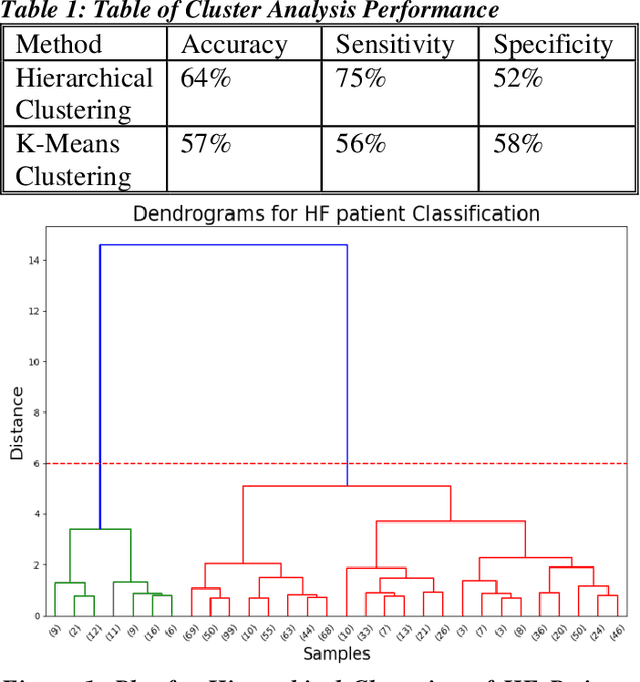
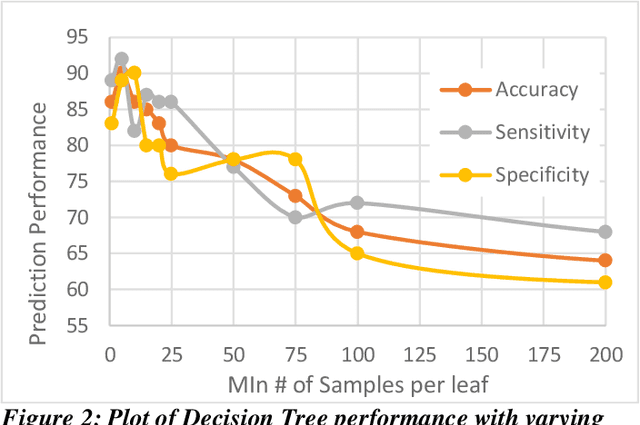
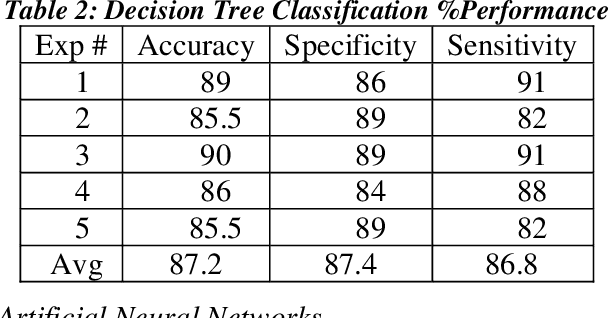
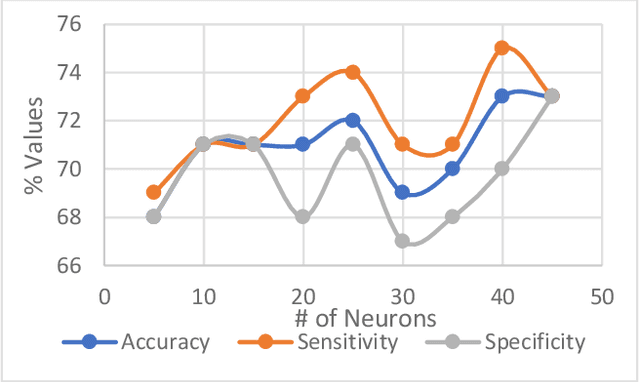
Abstract:Heart failure (HF) is a leading cause of morbidity, mortality, and health care costs. Prolonged conduction through the myocardium can occur with HF, and a device-driven approach, termed cardiac resynchronization therapy (CRT), can improve left ventricular (LV) myocardial conduction patterns. While a functional benefit of CRT has been demonstrated, a large proportion of HF patients (30-50%) receiving CRT do not show sufficient improvement. Moreover, identifying HF patients that would benefit from CRT prospectively remains a clinical challenge. Accordingly, strategies to effectively predict those HF patients that would derive a functional benefit from CRT holds great medical and socio-economic importance. Thus, we used machine learning methods of classifying HF patients, namely Cluster Analysis, Decision Trees, and Artificial neural networks, to develop predictive models of individual outcomes following CRT. Clinical, functional, and biomarker data were collected in HF patients before and following CRT. A prospective 6-month endpoint of a reduction in LV volume was defined as a CRT response. Using this approach (418 responders, 412 non-responders), each with 56 parameters, we could classify HF patients based on their response to CRT with more than 95% success. We have demonstrated that using machine learning approaches can identify HF patients with a high probability of a positive CRT response (95% accuracy), and of equal importance, identify those HF patients that would not derive a functional benefit from CRT. Developing this approach into a clinical algorithm to assist in clinical decision-making regarding the use of CRT in HF patients would potentially improve outcomes and reduce health care costs.
MedSensor: Medication Adherence Monitoring Using Neural Networks on Smartwatch Accelerometer Sensor Data
May 19, 2021

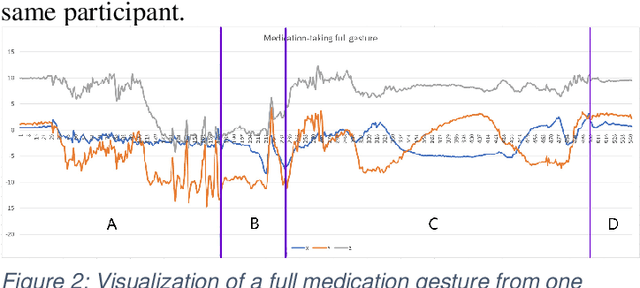

Abstract:Poor medication adherence presents serious economic and health problems including compromised treatment effectiveness, medical complications, and loss of billions of dollars in wasted medicine or procedures. Though various interventions have been proposed to address this problem, there is an urgent need to leverage light, smart, and minimally obtrusive technology such as smartwatches to develop user tools to improve medication use and adherence. In this study, we conducted several experiments on medication-taking activities, developed a smartwatch android application to collect the accelerometer hand gesture data from the smartwatch, and conveyed the data collected to a central cloud database. We developed neural networks, then trained the networks on the sensor data to recognize medication and non-medication gestures. With the proposed machine learning algorithm approach, this study was able to achieve average accuracy scores of 97% on the protocol-guided gesture data, and 95% on natural gesture data.
Reduction in the complexity of 1D 1H-NMR spectra by the use of Frequency to Information Transformation
Dec 16, 2020
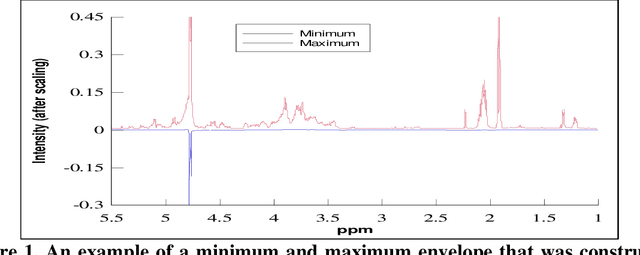
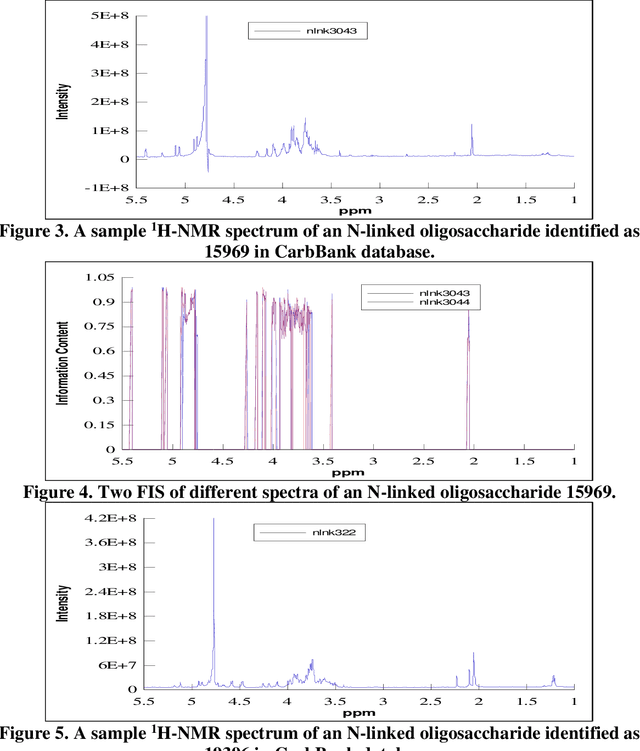
Abstract:Analysis of 1H-NMR spectra is often hindered by large variations that occur during the collection of these spectra. Large solvent and standard peaks, base line drift and negative peaks (due to improper phasing) are among some of these variations. Furthermore, some instrument dependent alterations, such as incorrect shimming, are also embedded in the recorded spectrum. The unpredictable nature of these alterations of the signal has rendered the automated and instrument independent computer analysis of these spectra unreliable. In this paper, a novel method of extracting the information content of a signal (in this paper, frequency domain 1H-NMR spectrum), called the frequency-information transformation (FIT), is presented and compared to a previously used method (SPUTNIK). FIT can successfully extract the relevant information to a pattern matching task present in a signal, while discarding the remainder of a signal by transforming a Fourier transformed signal into an information spectrum (IS). This technique exhibits the ability of decreasing the inter-class correlation coefficients while increasing the intra-class correlation coefficients. Different spectra of the same molecule, in other words, will resemble more to each other while the spectra of different molecules will look more different from each other. This feature allows easier automated identification and analysis of molecules based on their spectral signatures using computer algorithms.
TALI: Protein Structure Alignment Using Backbone Torsion Angles
Dec 12, 2020


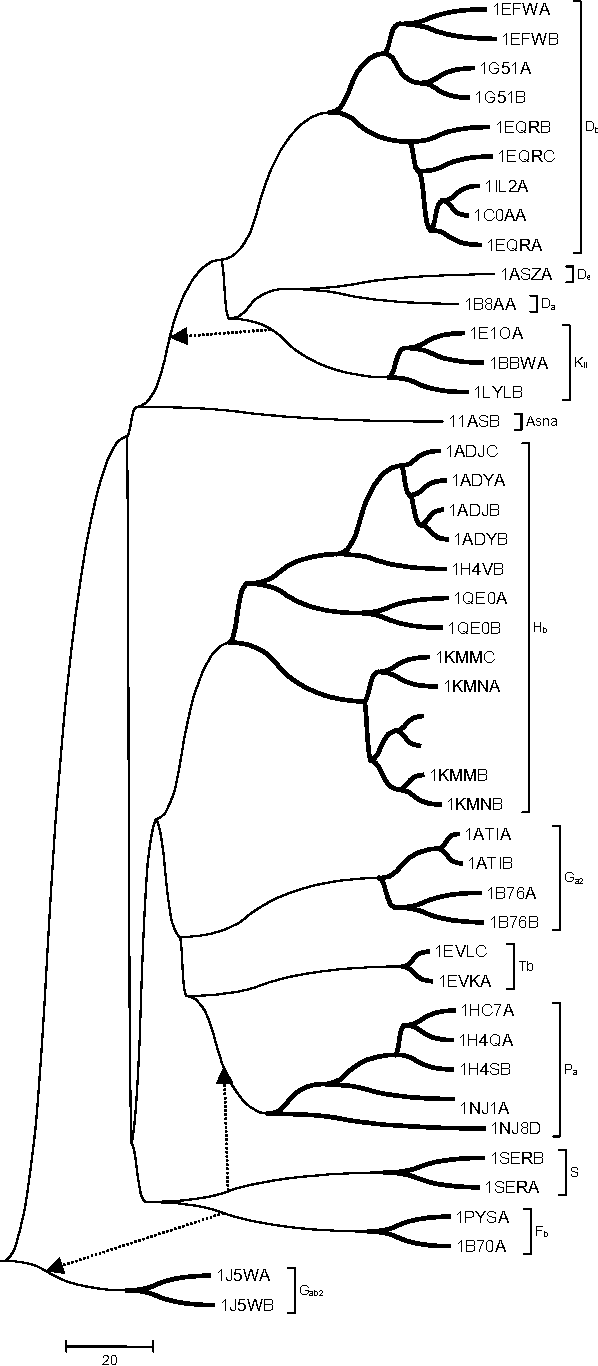
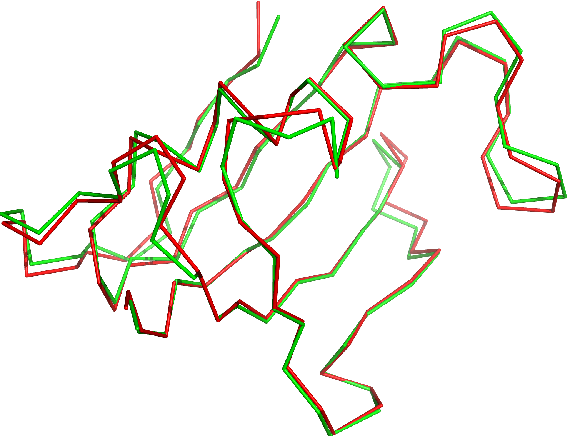
Abstract:This article introduces a novel protein structure alignment method (named TALI) based on the protein backbone torsion angle instead of the more traditional distance matrix. Because the structural alignment of the two proteins is based on the comparison of two sequences of numbers (backbone torsion angles), we can take advantage of a large number of well-developed methods such as Smith-Waterman or Needleman-Wunsch. Here we report the result of TALI in comparison to other structure alignment methods such as DALI, CE, and SSM ass well as sequence alignment based on PSI-BLAST. TALI demonstrated great success over all other methods in application to challenging proteins. TALI was more successful in recognizing remote structural homology. TALI also demonstrated an ability to identify structural homology between two proteins where the structural difference was due to a rotation of internal domains by nearly 180$^\circ$.
* Seven pages
An Investigation in Optimal Encoding of Protein Primary Sequence for Structure Prediction by Artificial Neural Networks
Aug 02, 2020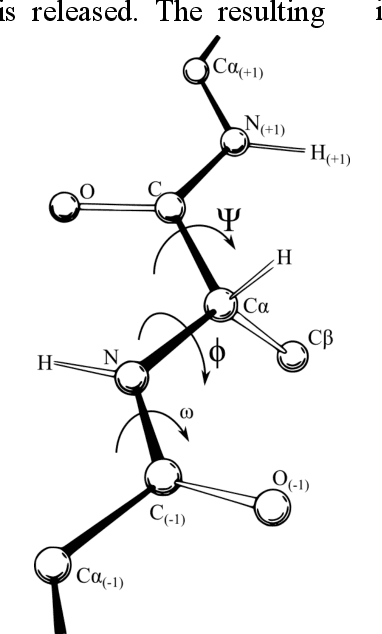

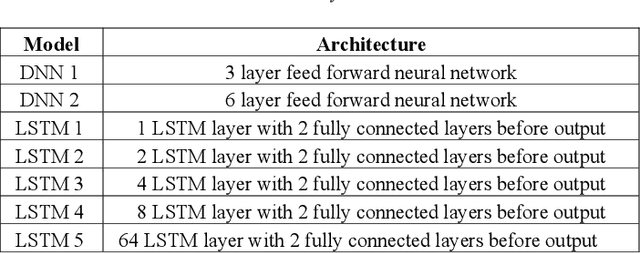
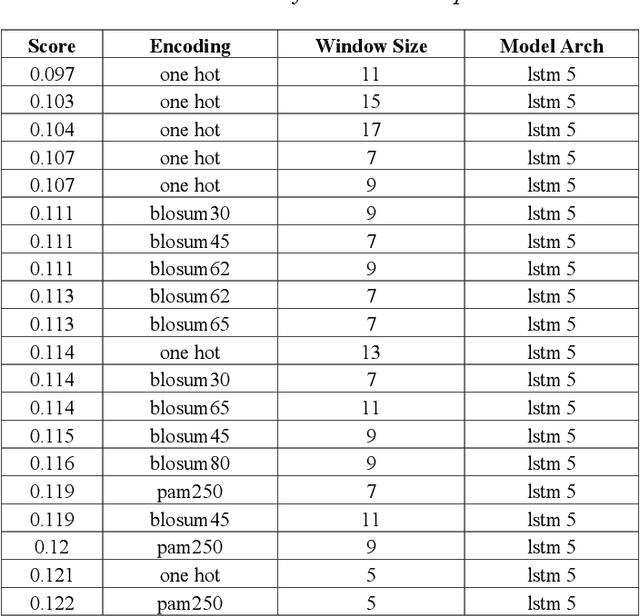
Abstract:Machine learning and the use of neural networks has increased precipitously over the past few years primarily due to the ever-increasing accessibility to data and the growth of computation power. It has become increasingly easy to harness the power of machine learning for predictive tasks. Protein structure prediction is one area where neural networks are becoming increasingly popular and successful. Although very powerful, the use of ANN require selection of most appropriate input/output encoding, architecture, and class to produce the optimal results. In this investigation we have explored and evaluated the effect of several conventional and newly proposed input encodings and selected an optimal architecture. We considered 11 variations of input encoding, 11 alternative window sizes, and 7 different architectures. In total, we evaluated 2,541 permutations in application to the training and testing of more than 10,000 protein structures over the course of 3 months. Our investigations concluded that one-hot encoding, the use of LSTMs, and window sizes of 9, 11, and 15 produce the optimal outcome. Through this optimization, we were able to improve the quality of protein structure prediction by predicting the {\phi} dihedrals to within 14{\deg} - 16{\deg} and {\psi} dihedrals to within 23{\deg}- 25{\deg}. This is a notable improvement compared to previously similar investigations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge