Weigang Cui
SUGAR: Spherical Ultrafast Graph Attention Framework for Cortical Surface Registration
Jul 02, 2023



Abstract:Cortical surface registration plays a crucial role in aligning cortical functional and anatomical features across individuals. However, conventional registration algorithms are computationally inefficient. Recently, learning-based registration algorithms have emerged as a promising solution, significantly improving processing efficiency. Nonetheless, there remains a gap in the development of a learning-based method that exceeds the state-of-the-art conventional methods simultaneously in computational efficiency, registration accuracy, and distortion control, despite the theoretically greater representational capabilities of deep learning approaches. To address the challenge, we present SUGAR, a unified unsupervised deep-learning framework for both rigid and non-rigid registration. SUGAR incorporates a U-Net-based spherical graph attention network and leverages the Euler angle representation for deformation. In addition to the similarity loss, we introduce fold and multiple distortion losses, to preserve topology and minimize various types of distortions. Furthermore, we propose a data augmentation strategy specifically tailored for spherical surface registration, enhancing the registration performance. Through extensive evaluation involving over 10,000 scans from 7 diverse datasets, we showed that our framework exhibits comparable or superior registration performance in accuracy, distortion, and test-retest reliability compared to conventional and learning-based methods. Additionally, SUGAR achieves remarkable sub-second processing times, offering a notable speed-up of approximately 12,000 times in registering 9,000 subjects from the UK Biobank dataset in just 32 minutes. This combination of high registration performance and accelerated processing time may greatly benefit large-scale neuroimaging studies.
Boosted Convolutional Neural Networks for Motor Imagery EEG Decoding with Multiwavelet-based Time-Frequency Conditional Granger Causality Analysis
Oct 22, 2018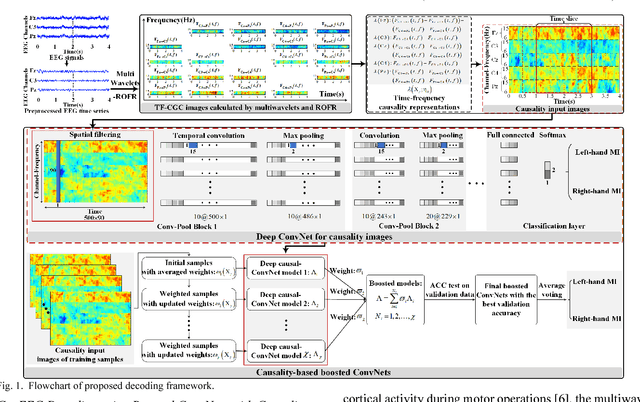
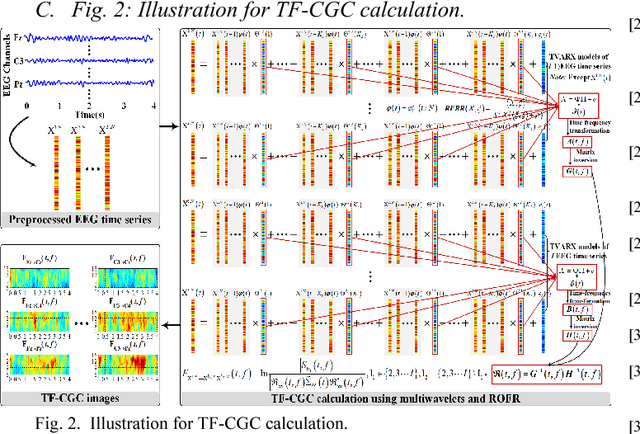
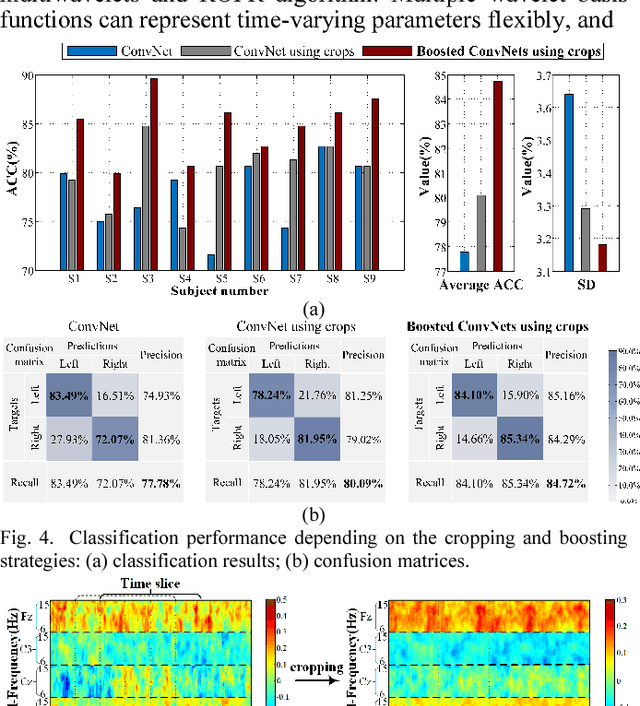
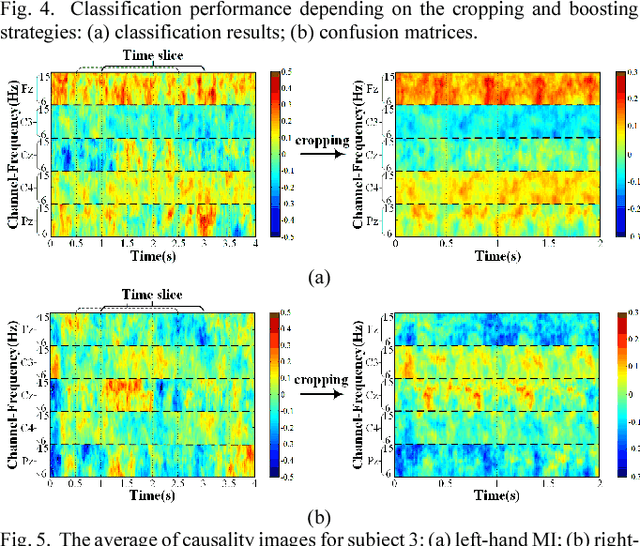
Abstract:Decoding EEG signals of different mental states is a challenging task for brain-computer interfaces (BCIs) due to nonstationarity of perceptual decision processes. This paper presents a novel boosted convolutional neural networks (ConvNets) decoding scheme for motor imagery (MI) EEG signals assisted by the multiwavelet-based time-frequency (TF) causality analysis. Specifically, multiwavelet basis functions are first combined with Geweke spectral measure to obtain high-resolution TF-conditional Granger causality (CGC) representations, where a regularized orthogonal forward regression (ROFR) algorithm is adopted to detect a parsimonious model with good generalization performance. The causality images for network input preserving time, frequency and location information of connectivity are then designed based on the TF-CGC distributions of alpha band multichannel EEG signals. Further constructed boosted ConvNets by using spatio-temporal convolutions as well as advances in deep learning including cropping and boosting methods, to extract discriminative causality features and classify MI tasks. Our proposed approach outperforms the competition winner algorithm with 12.15% increase in average accuracy and 74.02% decrease in associated inter subject standard deviation for the same binary classification on BCI competition-IV dataset-IIa. Experiment results indicate that the boosted ConvNets with causality images works well in decoding MI-EEG signals and provides a promising framework for developing MI-BCI systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge