Veysel Kocaman
Beyond Accuracy: Automated De-Identification of Large Real-World Clinical Text Datasets
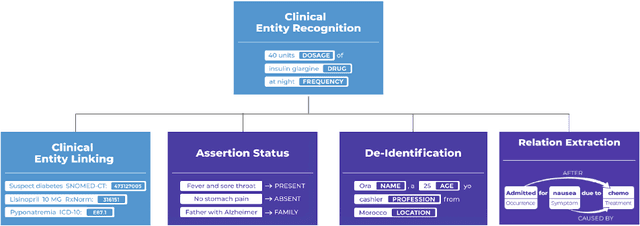
Dec 13, 2023Abstract:Recent research advances achieve human-level accuracy for de-identifying free-text clinical notes on research datasets, but gaps remain in reproducing this in large real-world settings. This paper summarizes lessons learned from building a system used to de-identify over one billion real clinical notes, in a fully automated way, that was independently certified by multiple organizations for production use. A fully automated solution requires a very high level of accuracy that does not require manual review. A hybrid context-based model architecture is described, which outperforms a Named Entity Recogniton (NER) - only model by 10% on the i2b2-2014 benchmark. The proposed system makes 50%, 475%, and 575% fewer errors than the comparable AWS, Azure, and GCP services respectively while also outperforming ChatGPT by 33%. It exceeds 98% coverage of sensitive data across 7 European languages, without a need for fine tuning. A second set of described models enable data obfuscation -- replacing sensitive data with random surrogates -- while retaining name, date, gender, clinical, and format consistency. Both the practical need and the solution architecture that provides for reliable & linked anonymized documents are described.
Saliency Can Be All You Need In Contrastive Self-Supervised Learning
Oct 30, 2022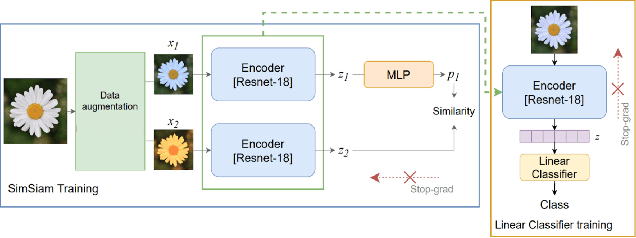

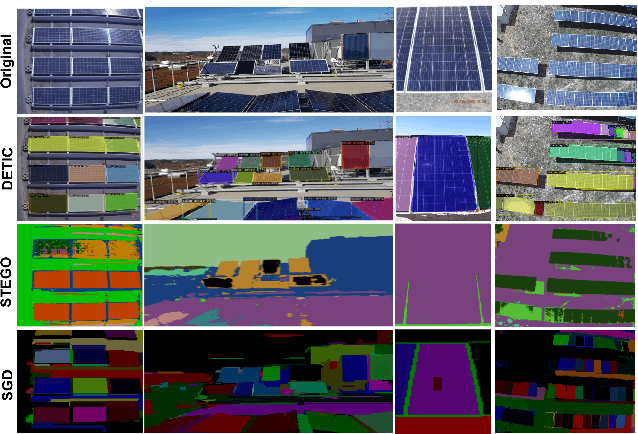
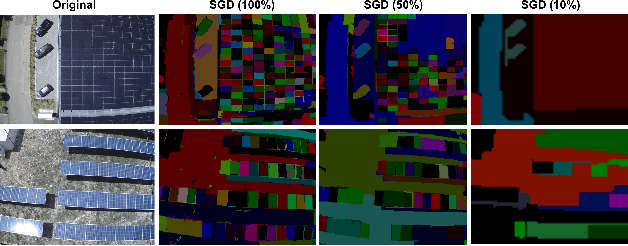
Abstract:We propose an augmentation policy for Contrastive Self-Supervised Learning (SSL) in the form of an already established Salient Image Segmentation technique entitled Global Contrast based Salient Region Detection. This detection technique, which had been devised for unrelated Computer Vision tasks, was empirically observed to play the role of an augmentation facilitator within the SSL protocol. This observation is rooted in our practical attempts to learn, by SSL-fashion, aerial imagery of solar panels, which exhibit challenging boundary patterns. Upon the successful integration of this technique on our problem domain, we formulated a generalized procedure and conducted a comprehensive, systematic performance assessment with various Contrastive SSL algorithms subject to standard augmentation techniques. This evaluation, which was conducted across multiple datasets, indicated that the proposed technique indeed contributes to SSL. We hypothesize whether salient image segmentation may suffice as the only augmentation policy in Contrastive SSL when treating downstream segmentation tasks.
Understanding COVID-19 News Coverage using Medical NLP
Mar 19, 2022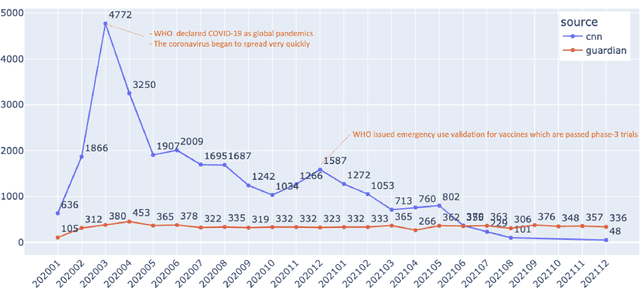
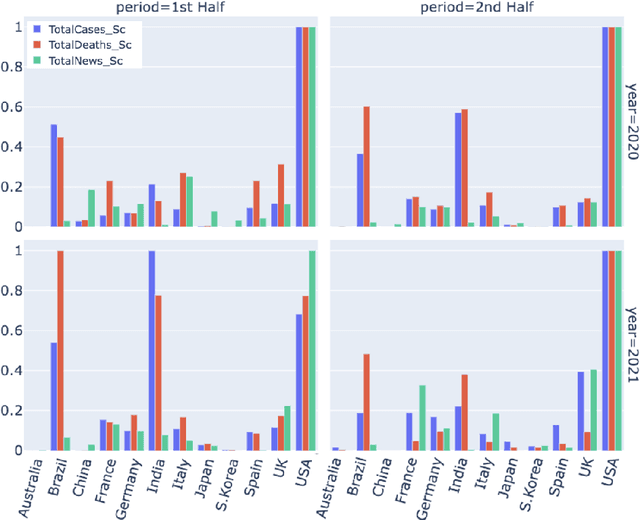
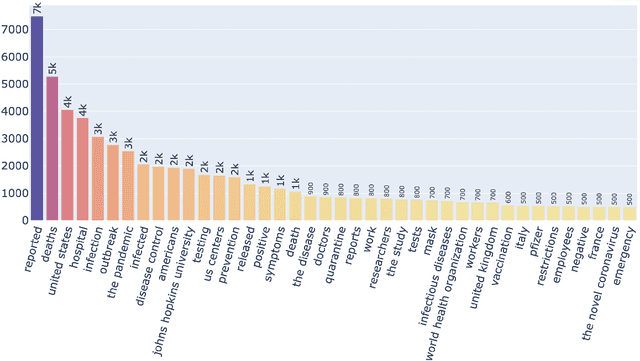
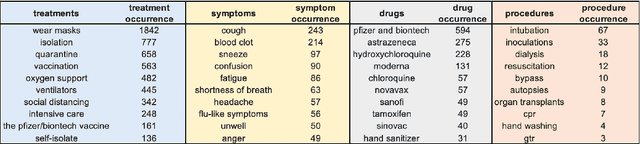
Abstract:Being a global pandemic, the COVID-19 outbreak received global media attention. In this study, we analyze news publications from CNN and The Guardian - two of the world's most influential media organizations. The dataset includes more than 36,000 articles, analyzed using the clinical and biomedical Natural Language Processing (NLP) models from the Spark NLP for Healthcare library, which enables a deeper analysis of medical concepts than previously achieved. The analysis covers key entities and phrases, observed biases, and change over time in news coverage by correlating mined medical symptoms, procedures, drugs, and guidance with commonly mentioned demographic and occupational groups. Another analysis is of extracted Adverse Drug Events about drug and vaccine manufacturers, which when reported by major news outlets has an impact on vaccine hesitancy.
Mining Adverse Drug Reactions from Unstructured Mediums at Scale
Jan 06, 2022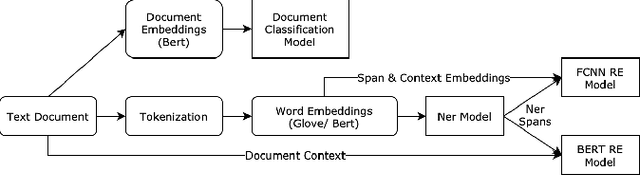
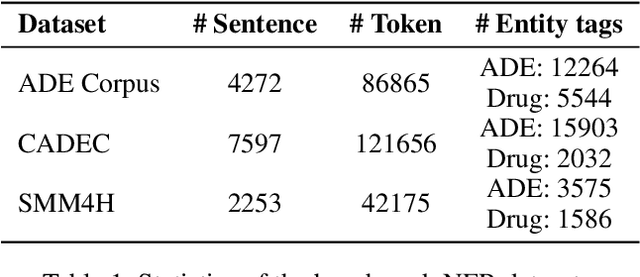


Abstract:Adverse drug reactions / events (ADR/ADE) have a major impact on patient health and health care costs. Detecting ADR's as early as possible and sharing them with regulators, pharma companies, and healthcare providers can prevent morbidity and save many lives. While most ADR's are not reported via formal channels, they are often documented in a variety of unstructured conversations such as social media posts by patients, customer support call transcripts, or CRM notes of meetings between healthcare providers and pharma sales reps. In this paper, we propose a natural language processing (NLP) solution that detects ADR's in such unstructured free-text conversations, which improves on previous work in three ways. First, a new Named Entity Recognition (NER) model obtains new state-of-the-art accuracy for ADR and Drug entity extraction on the ADE, CADEC, and SMM4H benchmark datasets (91.75%, 78.76%, and 83.41% F1 scores respectively). Second, two new Relation Extraction (RE) models are introduced - one based on BioBERT while the other utilizing crafted features over a Fully Connected Neural Network (FCNN) - are shown to perform on par with existing state-of-the-art models, and outperform them when trained with a supplementary clinician-annotated RE dataset. Third, a new text classification model, for deciding if a conversation includes an ADR, obtains new state-of-the-art accuracy on the CADEC dataset (86.69% F1 score). The complete solution is implemented as a unified NLP pipeline in a production-grade library built on top of Apache Spark, making it natively scalable and able to process millions of batch or streaming records on commodity clusters.
Deeper Clinical Document Understanding Using Relation Extraction
Dec 25, 2021
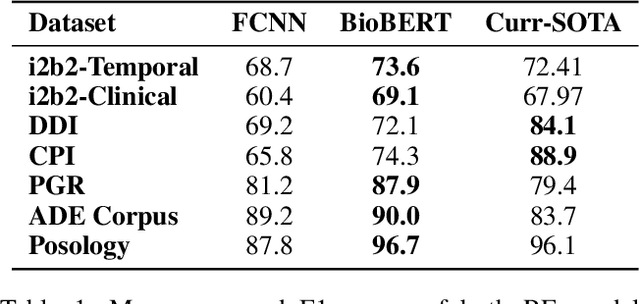
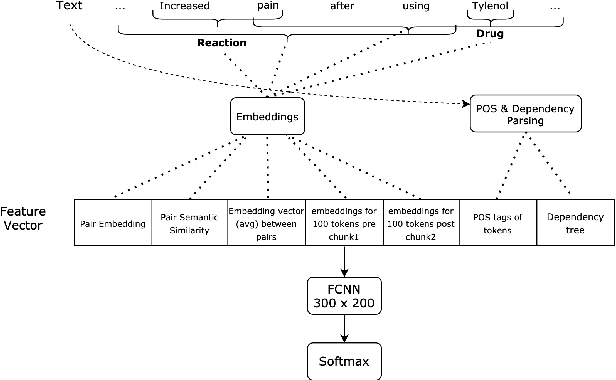

Abstract:The surging amount of biomedical literature & digital clinical records presents a growing need for text mining techniques that can not only identify but also semantically relate entities in unstructured data. In this paper we propose a text mining framework comprising of Named Entity Recognition (NER) and Relation Extraction (RE) models, which expands on previous work in three main ways. First, we introduce two new RE model architectures -- an accuracy-optimized one based on BioBERT and a speed-optimized one utilizing crafted features over a Fully Connected Neural Network (FCNN). Second, we evaluate both models on public benchmark datasets and obtain new state-of-the-art F1 scores on the 2012 i2b2 Clinical Temporal Relations challenge (F1 of 73.6, +1.2% over the previous SOTA), the 2010 i2b2 Clinical Relations challenge (F1 of 69.1, +1.2%), the 2019 Phenotype-Gene Relations dataset (F1 of 87.9, +8.5%), the 2012 Adverse Drug Events Drug-Reaction dataset (F1 of 90.0, +6.3%), and the 2018 n2c2 Posology Relations dataset (F1 of 96.7, +0.6%). Third, we show two practical applications of this framework -- for building a biomedical knowledge graph and for improving the accuracy of mapping entities to clinical codes. The system is built using the Spark NLP library which provides a production-grade, natively scalable, hardware-optimized, trainable & tunable NLP framework.
The Unreasonable Effectiveness of the Final Batch Normalization Layer
Sep 18, 2021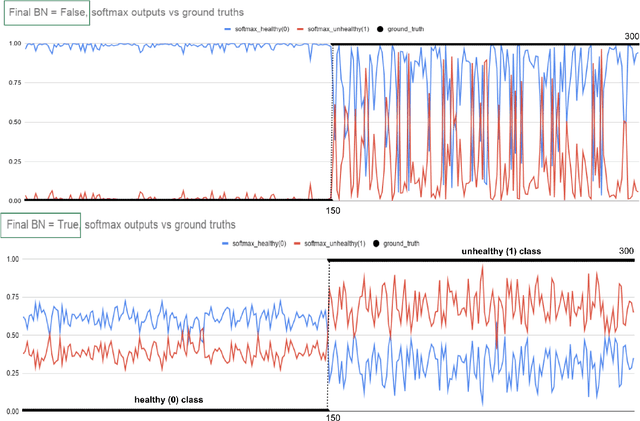

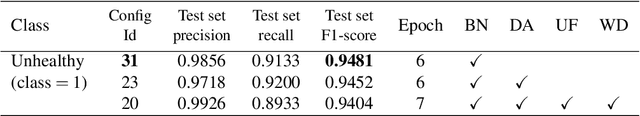
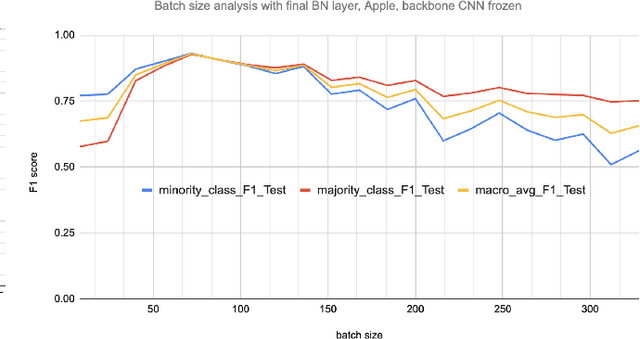
Abstract:Early-stage disease indications are rarely recorded in real-world domains, such as Agriculture and Healthcare, and yet, their accurate identification is critical in that point of time. In this type of highly imbalanced classification problems, which encompass complex features, deep learning (DL) is much needed because of its strong detection capabilities. At the same time, DL is observed in practice to favor majority over minority classes and consequently suffer from inaccurate detection of the targeted early-stage indications. In this work, we extend the study done by Kocaman et al., 2020, showing that the final BN layer, when placed before the softmax output layer, has a considerable impact in highly imbalanced image classification problems as well as undermines the role of the softmax outputs as an uncertainty measure. This current study addresses additional hypotheses and reports on the following findings: (i) the performance gain after adding the final BN layer in highly imbalanced settings could still be achieved after removing this additional BN layer in inference; (ii) there is a certain threshold for the imbalance ratio upon which the progress gained by the final BN layer reaches its peak; (iii) the batch size also plays a role and affects the outcome of the final BN application; (iv) the impact of the BN application is also reproducible on other datasets and when utilizing much simpler neural architectures; (v) the reported BN effect occurs only per a single majority class and multiple minority classes i.e., no improvements are evident when there are two majority classes; and finally, (vi) utilizing this BN layer with sigmoid activation has almost no impact when dealing with a strongly imbalanced image classification tasks.
Spark NLP: Natural Language Understanding at Scale
Jan 26, 2021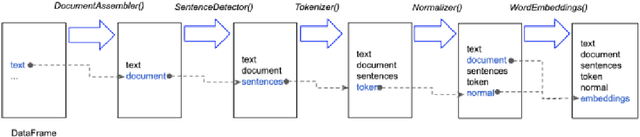
Abstract:Spark NLP is a Natural Language Processing (NLP) library built on top of Apache Spark ML. It provides simple, performant and accurate NLP annotations for machine learning pipelines that can scale easily in a distributed environment. Spark NLP comes with 1100 pre trained pipelines and models in more than 192 languages. It supports nearly all the NLP tasks and modules that can be used seamlessly in a cluster. Downloaded more than 2.7 million times and experiencing nine times growth since January 2020, Spark NLP is used by 54% of healthcare organizations as the worlds most widely used NLP library in the enterprise.
Improving Clinical Document Understanding on COVID-19 Research with Spark NLP
Dec 07, 2020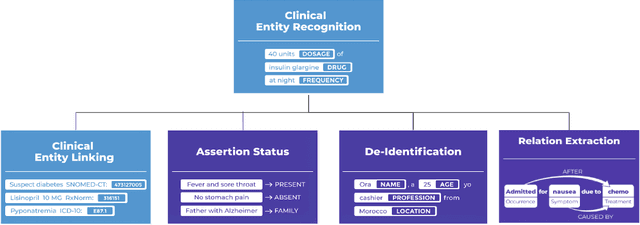
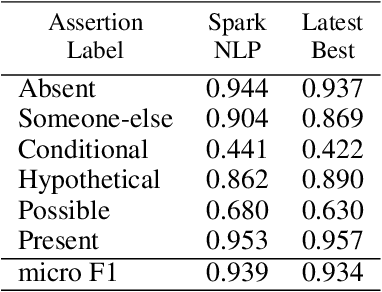
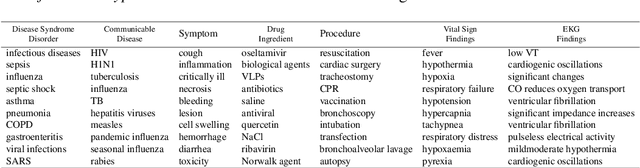
Abstract:Following the global COVID-19 pandemic, the number of scientific papers studying the virus has grown massively, leading to increased interest in automated literate review. We present a clinical text mining system that improves on previous efforts in three ways. First, it can recognize over 100 different entity types including social determinants of health, anatomy, risk factors, and adverse events in addition to other commonly used clinical and biomedical entities. Second, the text processing pipeline includes assertion status detection, to distinguish between clinical facts that are present, absent, conditional, or about someone other than the patient. Third, the deep learning models used are more accurate than previously available, leveraging an integrated pipeline of state-of-the-art pretrained named entity recognition models, and improving on the previous best performing benchmarks for assertion status detection. We illustrate extracting trends and insights, e.g. most frequent disorders and symptoms, and most common vital signs and EKG findings, from the COVID-19 Open Research Dataset (CORD-19). The system is built using the Spark NLP library which natively supports scaling to use distributed clusters, leveraging GPUs, configurable and reusable NLP pipelines, healthcare specific embeddings, and the ability to train models to support new entity types or human languages with no code changes.
Improving Model Accuracy for Imbalanced Image Classification Tasks by Adding a Final Batch Normalization Layer: An Empirical Study
Nov 12, 2020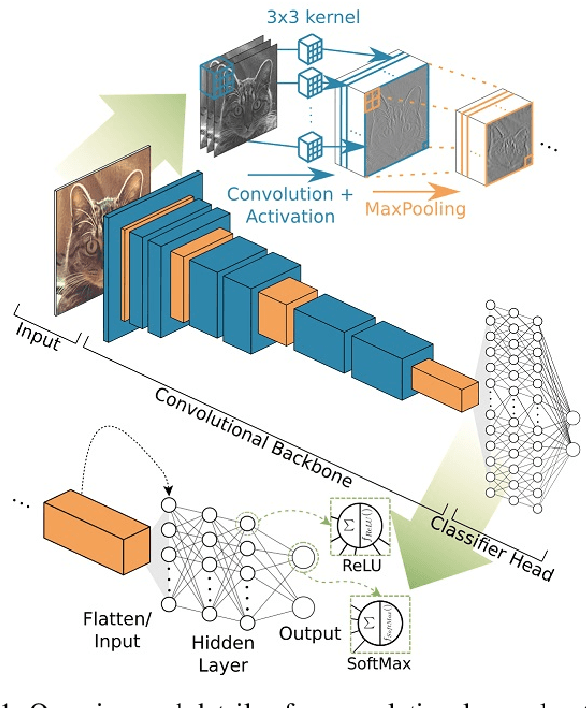
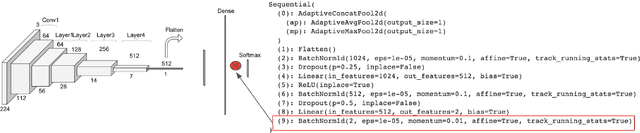
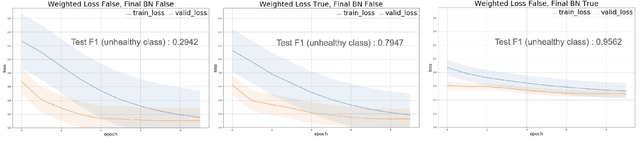
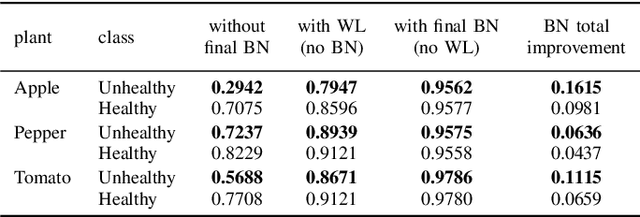
Abstract:Some real-world domains, such as Agriculture and Healthcare, comprise early-stage disease indications whose recording constitutes a rare event, and yet, whose precise detection at that stage is critical. In this type of highly imbalanced classification problems, which encompass complex features, deep learning (DL) is much needed because of its strong detection capabilities. At the same time, DL is observed in practice to favor majority over minority classes and consequently suffer from inaccurate detection of the targeted early-stage indications. To simulate such scenarios, we artificially generate skewness (99% vs. 1%) for certain plant types out of the PlantVillage dataset as a basis for classification of scarce visual cues through transfer learning. By randomly and unevenly picking healthy and unhealthy samples from certain plant types to form a training set, we consider a base experiment as fine-tuning ResNet34 and VGG19 architectures and then testing the model performance on a balanced dataset of healthy and unhealthy images. We empirically observe that the initial F1 test score jumps from 0.29 to 0.95 for the minority class upon adding a final Batch Normalization (BN) layer just before the output layer in VGG19. We demonstrate that utilizing an additional BN layer before the output layer in modern CNN architectures has a considerable impact in terms of minimizing the training time and testing error for minority classes in highly imbalanced data sets. Moreover, when the final BN is employed, minimizing the loss function may not be the best way to assure a high F1 test score for minority classes in such problems. That is, the network might perform better even if it is not confident enough while making a prediction; leading to another discussion about why softmax output is not a good uncertainty measure for DL models.
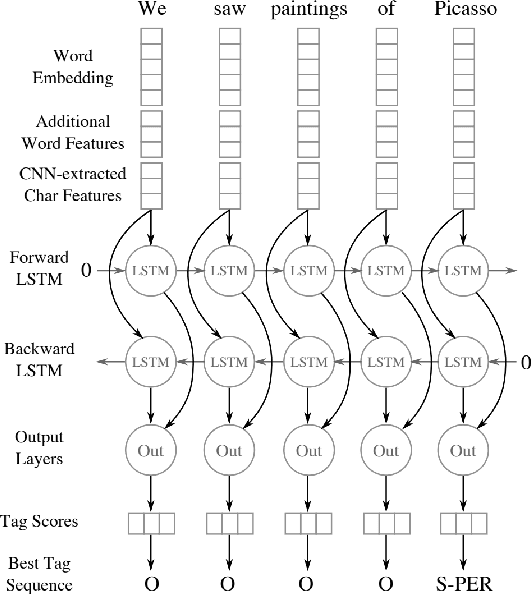
Biomedical Named Entity Recognition at Scale
Nov 12, 2020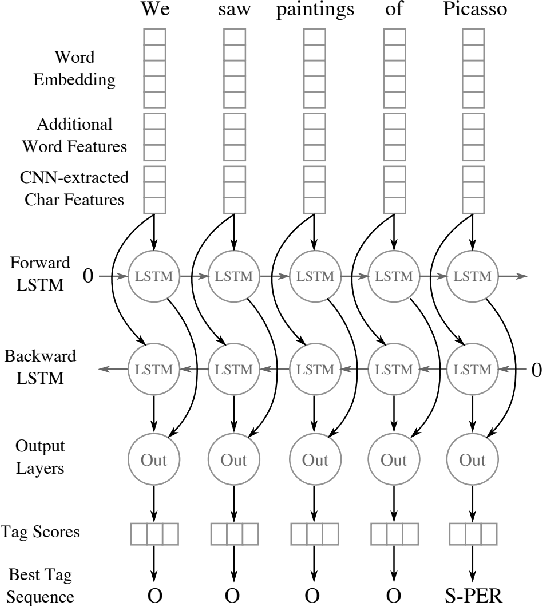
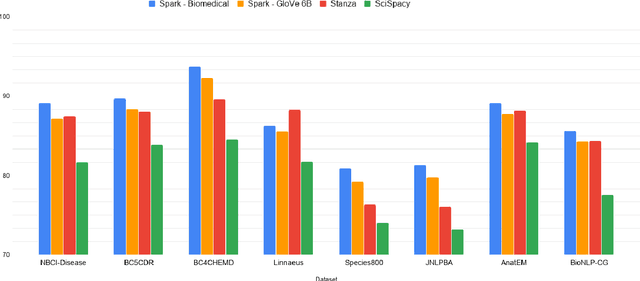
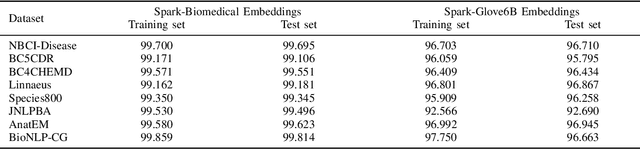
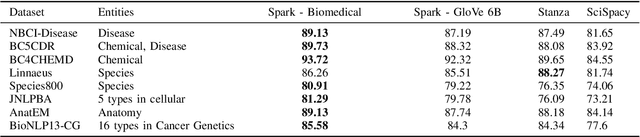
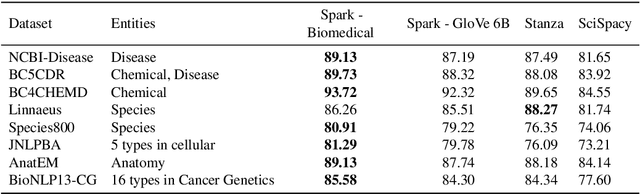
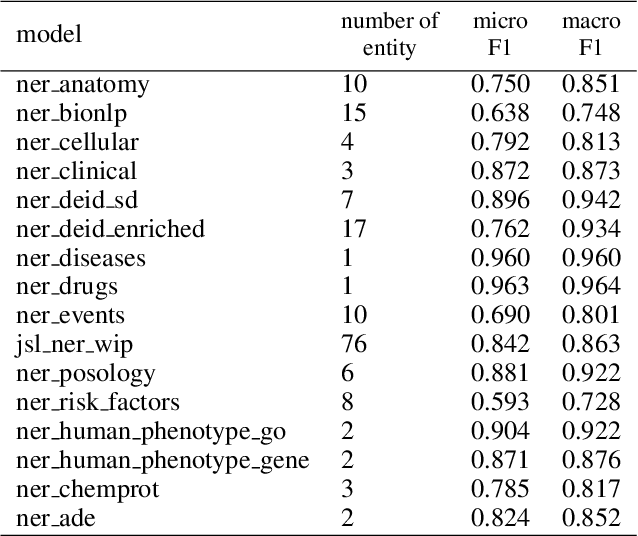
Abstract:Named entity recognition (NER) is a widely applicable natural language processing task and building block of question answering, topic modeling, information retrieval, etc. In the medical domain, NER plays a crucial role by extracting meaningful chunks from clinical notes and reports, which are then fed to downstream tasks like assertion status detection, entity resolution, relation extraction, and de-identification. Reimplementing a Bi-LSTM-CNN-Char deep learning architecture on top of Apache Spark, we present a single trainable NER model that obtains new state-of-the-art results on seven public biomedical benchmarks without using heavy contextual embeddings like BERT. This includes improving BC4CHEMD to 93.72% (4.1% gain), Species800 to 80.91% (4.6% gain), and JNLPBA to 81.29% (5.2% gain). In addition, this model is freely available within a production-grade code base as part of the open-source Spark NLP library; can scale up for training and inference in any Spark cluster; has GPU support and libraries for popular programming languages such as Python, R, Scala and Java; and can be extended to support other human languages with no code changes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge