Hasham Ul Haq
Beyond Accuracy: Automated De-Identification of Large Real-World Clinical Text Datasets
Dec 13, 2023Abstract:Recent research advances achieve human-level accuracy for de-identifying free-text clinical notes on research datasets, but gaps remain in reproducing this in large real-world settings. This paper summarizes lessons learned from building a system used to de-identify over one billion real clinical notes, in a fully automated way, that was independently certified by multiple organizations for production use. A fully automated solution requires a very high level of accuracy that does not require manual review. A hybrid context-based model architecture is described, which outperforms a Named Entity Recogniton (NER) - only model by 10% on the i2b2-2014 benchmark. The proposed system makes 50%, 475%, and 575% fewer errors than the comparable AWS, Azure, and GCP services respectively while also outperforming ChatGPT by 33%. It exceeds 98% coverage of sensitive data across 7 European languages, without a need for fine tuning. A second set of described models enable data obfuscation -- replacing sensitive data with random surrogates -- while retaining name, date, gender, clinical, and format consistency. Both the practical need and the solution architecture that provides for reliable & linked anonymized documents are described.
Understanding COVID-19 News Coverage using Medical NLP
Mar 19, 2022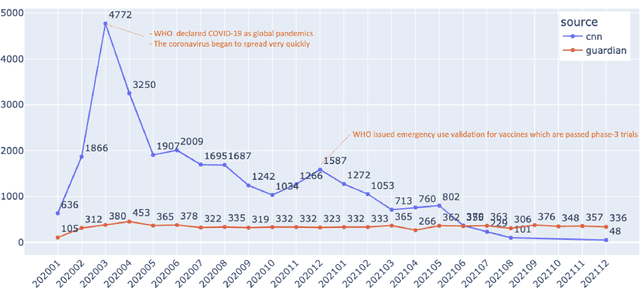
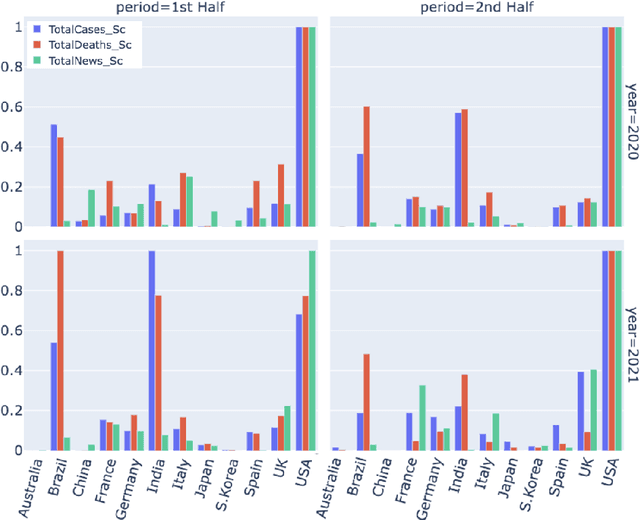
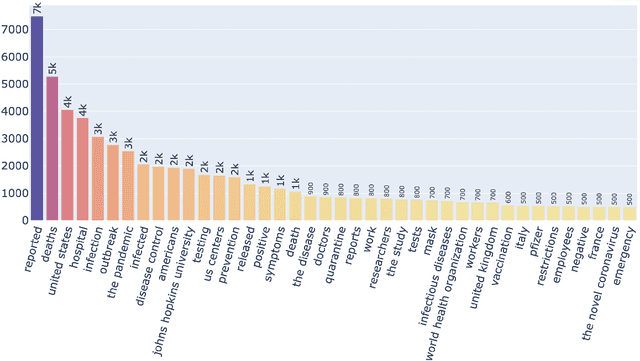
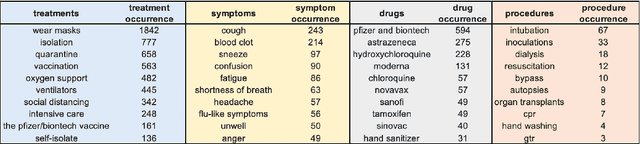
Abstract:Being a global pandemic, the COVID-19 outbreak received global media attention. In this study, we analyze news publications from CNN and The Guardian - two of the world's most influential media organizations. The dataset includes more than 36,000 articles, analyzed using the clinical and biomedical Natural Language Processing (NLP) models from the Spark NLP for Healthcare library, which enables a deeper analysis of medical concepts than previously achieved. The analysis covers key entities and phrases, observed biases, and change over time in news coverage by correlating mined medical symptoms, procedures, drugs, and guidance with commonly mentioned demographic and occupational groups. Another analysis is of extracted Adverse Drug Events about drug and vaccine manufacturers, which when reported by major news outlets has an impact on vaccine hesitancy.
Mining Adverse Drug Reactions from Unstructured Mediums at Scale
Jan 06, 2022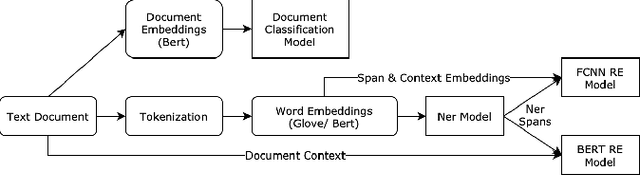
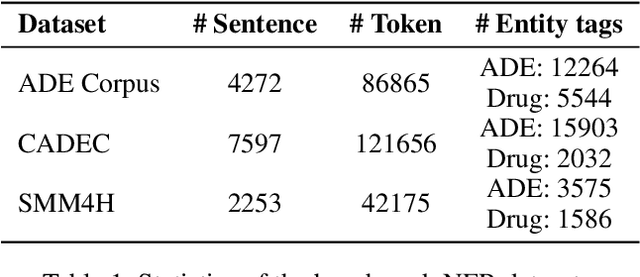


Abstract:Adverse drug reactions / events (ADR/ADE) have a major impact on patient health and health care costs. Detecting ADR's as early as possible and sharing them with regulators, pharma companies, and healthcare providers can prevent morbidity and save many lives. While most ADR's are not reported via formal channels, they are often documented in a variety of unstructured conversations such as social media posts by patients, customer support call transcripts, or CRM notes of meetings between healthcare providers and pharma sales reps. In this paper, we propose a natural language processing (NLP) solution that detects ADR's in such unstructured free-text conversations, which improves on previous work in three ways. First, a new Named Entity Recognition (NER) model obtains new state-of-the-art accuracy for ADR and Drug entity extraction on the ADE, CADEC, and SMM4H benchmark datasets (91.75%, 78.76%, and 83.41% F1 scores respectively). Second, two new Relation Extraction (RE) models are introduced - one based on BioBERT while the other utilizing crafted features over a Fully Connected Neural Network (FCNN) - are shown to perform on par with existing state-of-the-art models, and outperform them when trained with a supplementary clinician-annotated RE dataset. Third, a new text classification model, for deciding if a conversation includes an ADR, obtains new state-of-the-art accuracy on the CADEC dataset (86.69% F1 score). The complete solution is implemented as a unified NLP pipeline in a production-grade library built on top of Apache Spark, making it natively scalable and able to process millions of batch or streaming records on commodity clusters.
Deeper Clinical Document Understanding Using Relation Extraction
Dec 25, 2021
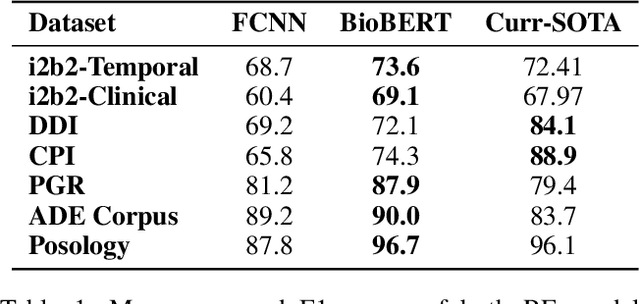
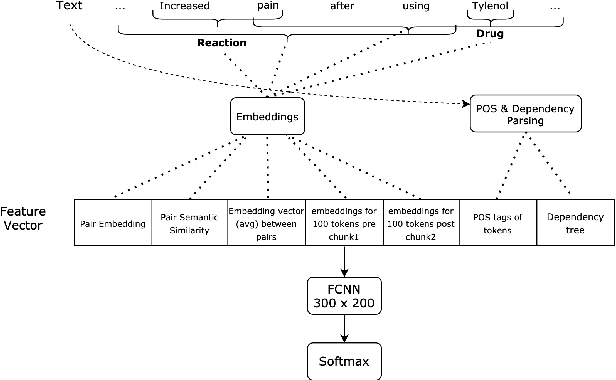

Abstract:The surging amount of biomedical literature & digital clinical records presents a growing need for text mining techniques that can not only identify but also semantically relate entities in unstructured data. In this paper we propose a text mining framework comprising of Named Entity Recognition (NER) and Relation Extraction (RE) models, which expands on previous work in three main ways. First, we introduce two new RE model architectures -- an accuracy-optimized one based on BioBERT and a speed-optimized one utilizing crafted features over a Fully Connected Neural Network (FCNN). Second, we evaluate both models on public benchmark datasets and obtain new state-of-the-art F1 scores on the 2012 i2b2 Clinical Temporal Relations challenge (F1 of 73.6, +1.2% over the previous SOTA), the 2010 i2b2 Clinical Relations challenge (F1 of 69.1, +1.2%), the 2019 Phenotype-Gene Relations dataset (F1 of 87.9, +8.5%), the 2012 Adverse Drug Events Drug-Reaction dataset (F1 of 90.0, +6.3%), and the 2018 n2c2 Posology Relations dataset (F1 of 96.7, +0.6%). Third, we show two practical applications of this framework -- for building a biomedical knowledge graph and for improving the accuracy of mapping entities to clinical codes. The system is built using the Spark NLP library which provides a production-grade, natively scalable, hardware-optimized, trainable & tunable NLP framework.
Intelligent EHRs: Predicting Procedure Codes From Diagnosis Codes
Dec 01, 2017

Abstract:In order to submit a claim to insurance companies, a doctor needs to code a patient encounter with both the diagnosis (ICDs) and procedures performed (CPTs) in an Electronic Health Record (EHR). Identifying and applying relevant procedures code is a cumbersome and time-consuming task as a doctor has to choose from around 13,000 procedure codes with no predefined one-to-one mapping. In this paper, we propose a state-of-the-art deep learning method for automatic and intelligent coding of procedures (CPTs) from the diagnosis codes (ICDs) entered by the doctor. Precisely, we cast the learning problem as a multi-label classification problem and use distributed representation to learn the input mapping of high-dimensional sparse ICDs codes. Our final model trained on 2.3 million claims is able to outperform existing rule-based probabilistic and association-rule mining based methods and has a recall of 90@3.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge