Deeper Clinical Document Understanding Using Relation Extraction
Paper and Code
Dec 25, 2021
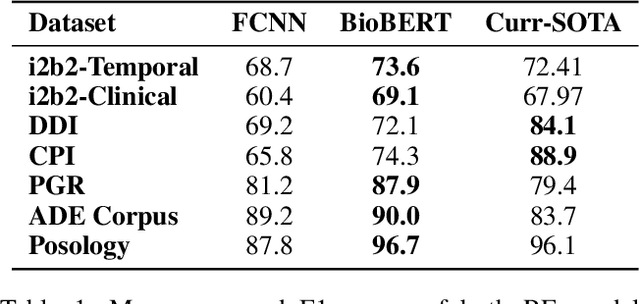
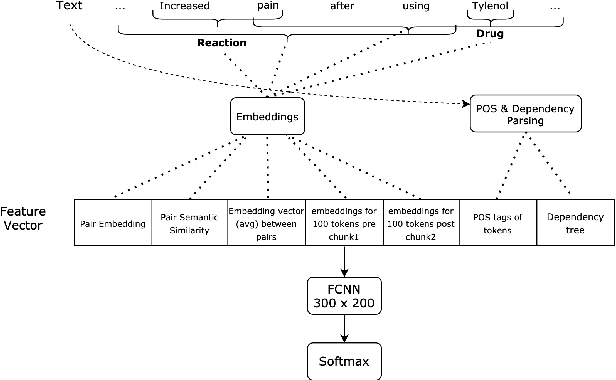

The surging amount of biomedical literature & digital clinical records presents a growing need for text mining techniques that can not only identify but also semantically relate entities in unstructured data. In this paper we propose a text mining framework comprising of Named Entity Recognition (NER) and Relation Extraction (RE) models, which expands on previous work in three main ways. First, we introduce two new RE model architectures -- an accuracy-optimized one based on BioBERT and a speed-optimized one utilizing crafted features over a Fully Connected Neural Network (FCNN). Second, we evaluate both models on public benchmark datasets and obtain new state-of-the-art F1 scores on the 2012 i2b2 Clinical Temporal Relations challenge (F1 of 73.6, +1.2% over the previous SOTA), the 2010 i2b2 Clinical Relations challenge (F1 of 69.1, +1.2%), the 2019 Phenotype-Gene Relations dataset (F1 of 87.9, +8.5%), the 2012 Adverse Drug Events Drug-Reaction dataset (F1 of 90.0, +6.3%), and the 2018 n2c2 Posology Relations dataset (F1 of 96.7, +0.6%). Third, we show two practical applications of this framework -- for building a biomedical knowledge graph and for improving the accuracy of mapping entities to clinical codes. The system is built using the Spark NLP library which provides a production-grade, natively scalable, hardware-optimized, trainable & tunable NLP framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge