Valérie Burdin
Fully automated workflow for the design of patient-specific orthopaedic implants: application to total knee arthroplasty
Mar 25, 2024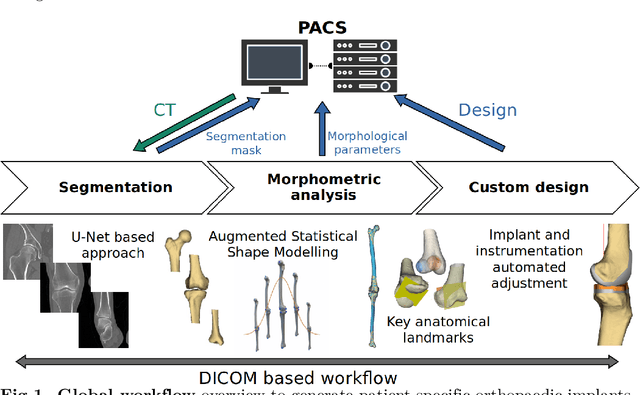
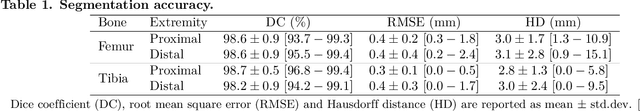
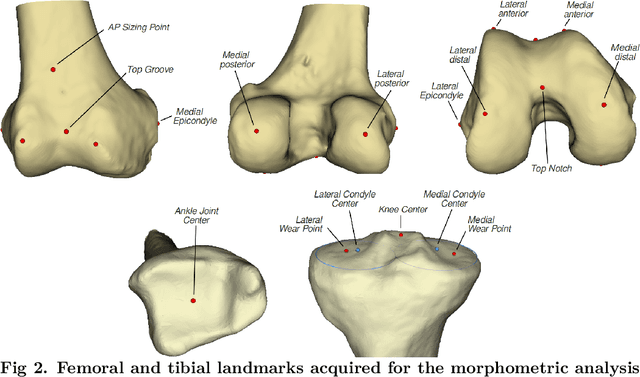
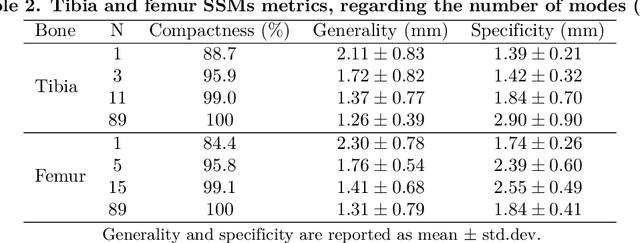
Abstract:Arthroplasty is commonly performed to treat joint osteoarthritis, reducing pain and improving mobility. While arthroplasty has known several technical improvements, a significant share of patients are still unsatisfied with their surgery. Personalised arthroplasty improves surgical outcomes however current solutions require delays, making it difficult to integrate in clinical routine. We propose a fully automated workflow to design patient-specific implants, presented for total knee arthroplasty, the most widely performed arthroplasty in the world nowadays. The proposed pipeline first uses artificial neural networks to segment the proximal and distal extremities of the femur and tibia. Then the full bones are reconstructed using augmented statistical shape models, combining shape and landmarks information. Finally, 77 morphological parameters are computed to design patient-specific implants. The developed workflow has been trained using 91 CT scans of lower limb and evaluated on 41 CT scans manually segmented, in terms of accuracy and execution time. The workflow accuracy was $0.4\pm0.2mm$ for the segmentation, $1.2\pm0.4mm$ for the full bones reconstruction, and $2.8\pm2.2mm$ for the anatomical landmarks determination. The custom implants fitted the patients' anatomy with $0.6\pm0.2mm$ accuracy. The whole process from segmentation to implants' design lasted about 5 minutes. The proposed workflow allows for a fast and reliable personalisation of knee implants, directly from the patient CT image without requiring any manual intervention. It establishes a patient-specific pre-operative planning for TKA in a very short time making it easily available for all patients. Combined with efficient implant manufacturing techniques, this solution could help answer the growing number of arthroplasties while reducing complications and improving the patients' satisfaction.
Generalizable multi-task, multi-domain deep segmentation of sparse pediatric imaging datasets via multi-scale contrastive regularization and multi-joint anatomical priors
Jul 27, 2022


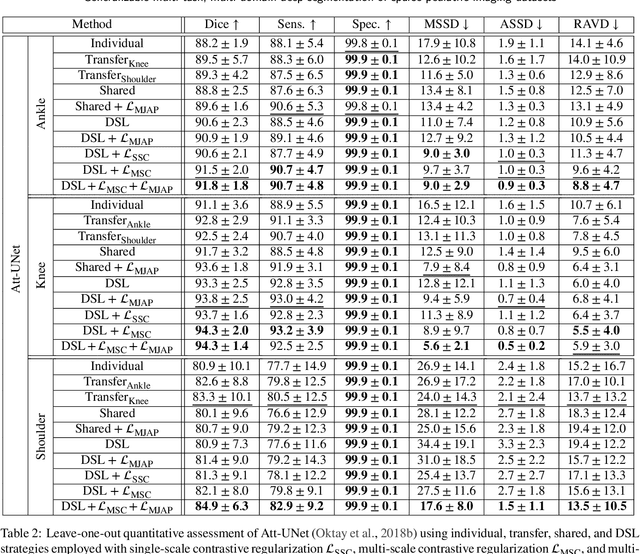
Abstract:Clinical diagnosis of the pediatric musculoskeletal system relies on the analysis of medical imaging examinations. In the medical image processing pipeline, semantic segmentation using deep learning algorithms enables an automatic generation of patient-specific three-dimensional anatomical models which are crucial for morphological evaluation. However, the scarcity of pediatric imaging resources may result in reduced accuracy and generalization performance of individual deep segmentation models. In this study, we propose to design a novel multi-task, multi-domain learning framework in which a single segmentation network is optimized over the union of multiple datasets arising from distinct parts of the anatomy. Unlike previous approaches, we simultaneously consider multiple intensity domains and segmentation tasks to overcome the inherent scarcity of pediatric data while leveraging shared features between imaging datasets. To further improve generalization capabilities, we employ a transfer learning scheme from natural image classification, along with a multi-scale contrastive regularization aimed at promoting domain-specific clusters in the shared representations, and multi-joint anatomical priors to enforce anatomically consistent predictions. We evaluate our contributions for performing bone segmentation using three scarce and pediatric imaging datasets of the ankle, knee, and shoulder joints. Our results demonstrate that the proposed approach outperforms individual, transfer, and shared segmentation schemes in Dice metric with statistically sufficient margins. The proposed model brings new perspectives towards intelligent use of imaging resources and better management of pediatric musculoskeletal disorders.
Anatomically Parameterized Statistical Shape Model: Explaining Morphometry through Statistical Learning
Feb 17, 2022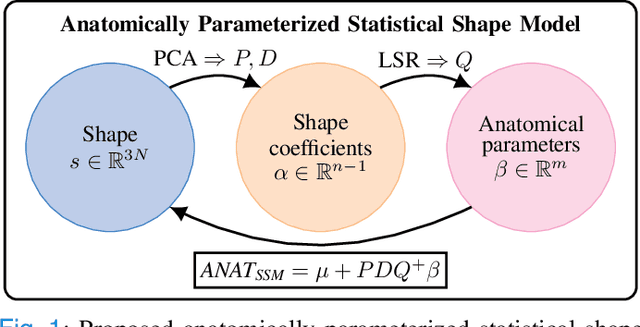
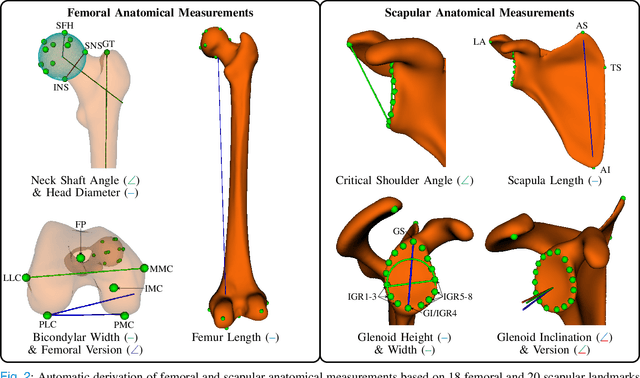
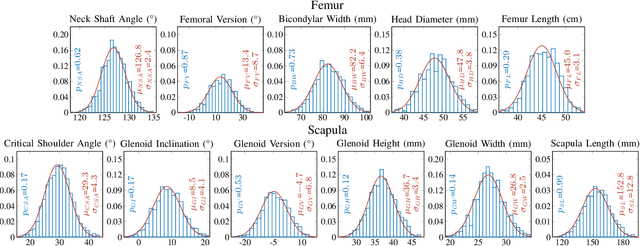
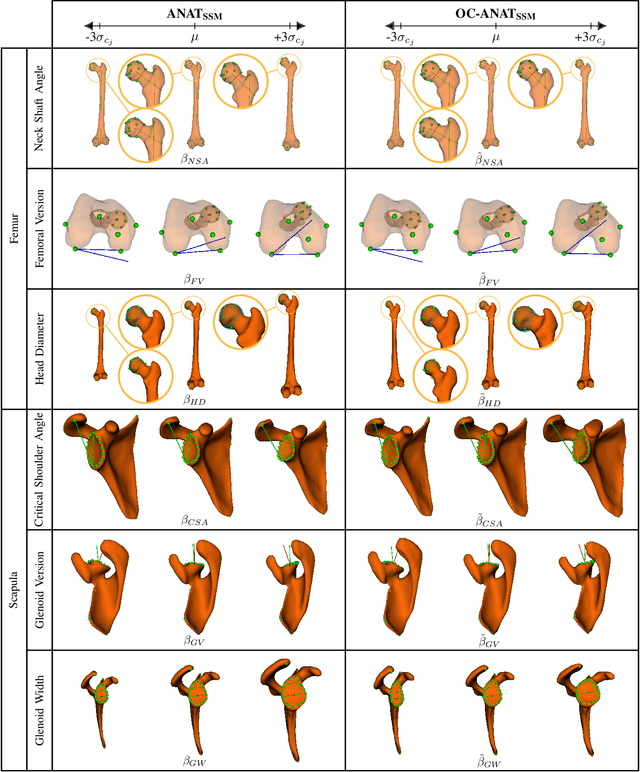
Abstract:Statistical shape models (SSMs) are a popular tool to conduct morphological analysis of anatomical structures which is a crucial step in clinical practices. However, shape representations through SSMs are based on shape coefficients and lack an explicit one-to-one relationship with anatomical measures of clinical relevance. While a shape coefficient embeds a combination of anatomical measures, a formalized approach to find the relationship between them remains elusive in the literature. This limits the use of SSMs to subjective evaluations in clinical practices. We propose a novel SSM controlled by anatomical parameters derived from morphometric analysis. The proposed anatomically parameterized SSM (ANAT-SSM) is based on learning a linear mapping between shape coefficients and selected anatomical parameters. This mapping is learned from a synthetic population generated by the standard SSM. Determining the pseudo-inverse of the mapping allows us to build the ANAT-SSM. We further impose orthogonality constraints to the anatomical parameterization to obtain independent shape variation patterns. The proposed contribution was evaluated on two skeletal databases of femoral and scapular bone shapes using clinically relevant anatomical parameters. Anatomical measures of the synthetically generated shapes exhibited realistic statistics. The learned matrices corroborated well with the obtained statistical relationship, while the two SSMs achieved moderate to excellent performance in predicting anatomical parameters on unseen shapes. This study demonstrates the use of anatomical representation for creating anatomically parameterized SSM and as a result, removes the limited clinical interpretability of standard SSMs. The proposed models could help analyze differences in relevant bone morphometry between populations, and be integrated in patient-specific pre-surgery planning or in-surgery assessment.
Dynamic multi feature-class Gaussian process models
Dec 08, 2021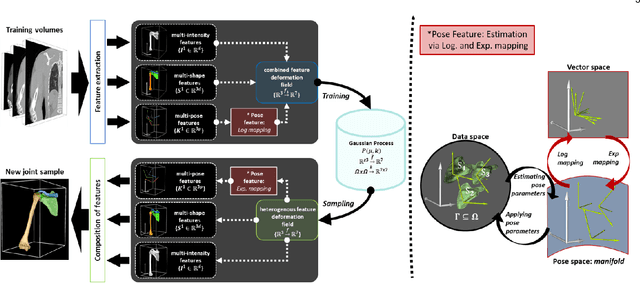
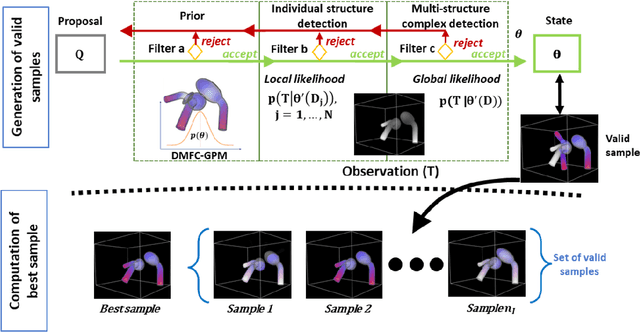
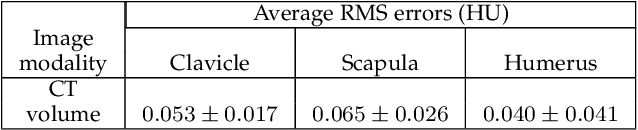
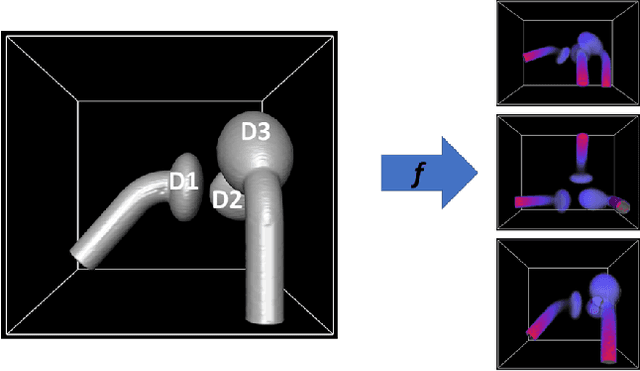
Abstract:In model-based medical image analysis, three features of interest are the shape of structures of interest, their relative pose, and image intensity profiles representative of some physical property. Often, these are modelled separately through statistical models by decomposing the object's features into a set of basis functions through principal geodesic analysis or principal component analysis. This study presents a statistical modelling method for automatic learning of shape, pose and intensity features in medical images which we call the Dynamic multi feature-class Gaussian process models (DMFC-GPM). A DMFC-GPM is a Gaussian process (GP)-based model with a shared latent space that encodes linear and non-linear variation. Our method is defined in a continuous domain with a principled way to represent shape, pose and intensity feature classes in a linear space, based on deformation fields. A deformation field-based metric is adapted in the method for modelling shape and intensity feature variation as well as for comparing rigid transformations (pose). Moreover, DMFC-GPMs inherit properties intrinsic to GPs including marginalisation and regression. Furthermore, they allow for adding additional pose feature variability on top of those obtained from the image acquisition process; what we term as permutation modelling. For image analysis tasks using DMFC-GPMs, we adapt Metropolis-Hastings algorithms making the prediction of features fully probabilistic. We validate the method using controlled synthetic data and we perform experiments on bone structures from CT images of the shoulder to illustrate the efficacy of the model for pose and shape feature prediction. The model performance results suggest that this new modelling paradigm is robust, accurate, accessible, and has potential applications including the management of musculoskeletal disorders and clinical decision making
Multi-Task, Multi-Domain Deep Segmentation with Shared Representations and Contrastive Regularization for Sparse Pediatric Datasets
May 21, 2021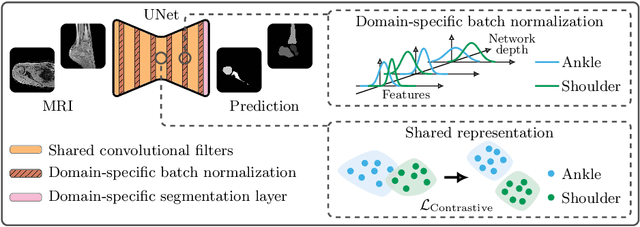



Abstract:Automatic segmentation of magnetic resonance (MR) images is crucial for morphological evaluation of the pediatric musculoskeletal system in clinical practice. However, the accuracy and generalization performance of individual segmentation models are limited due to the restricted amount of annotated pediatric data. Hence, we propose to train a segmentation model on multiple datasets, arising from different parts of the anatomy, in a multi-task and multi-domain learning framework. This approach allows to overcome the inherent scarcity of pediatric data while benefiting from a more robust shared representation. The proposed segmentation network comprises shared convolutional filters, domain-specific batch normalization parameters that compute the respective dataset statistics and a domain-specific segmentation layer. Furthermore, a supervised contrastive regularization is integrated to further improve generalization capabilities, by promoting intra-domain similarity and impose inter-domain margins in embedded space. We evaluate our contributions on two pediatric imaging datasets of the ankle and shoulder joints for bone segmentation. Results demonstrate that the proposed model outperforms state-of-the-art approaches.
Multi-Structure Deep Segmentation with Shape Priors and Latent Adversarial Regularization
Jan 25, 2021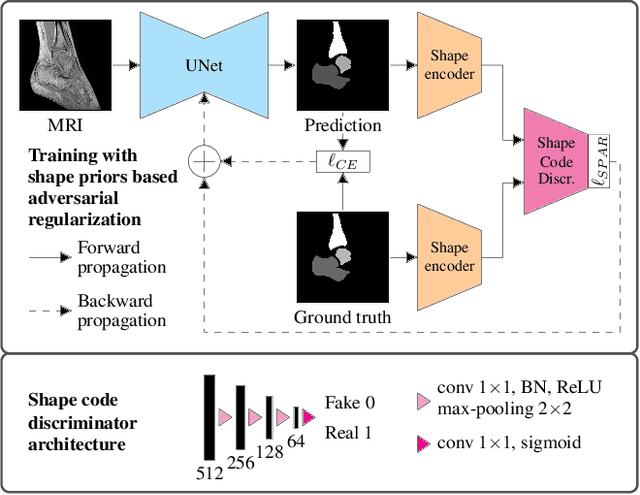


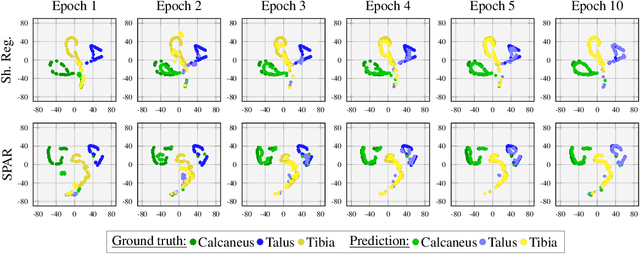
Abstract:Automatic segmentation of the musculoskeletal system in pediatric magnetic resonance (MR) images is a challenging but crucial task for morphological evaluation in clinical practice. We propose a deep learning-based regularized segmentation method for multi-structure bone delineation in MR images, designed to overcome the inherent scarcity and heterogeneity of pediatric data. Based on a newly devised shape code discriminator, our adversarial regularization scheme enforces the deep network to follow a learnt shape representation of the anatomy. The novel shape priors based adversarial regularization (SPAR) exploits latent shape codes arising from ground truth and predicted masks to guide the segmentation network towards more consistent and plausible predictions. Our contribution is compared to state-of-the-art regularization methods on two pediatric musculoskeletal imaging datasets from ankle and shoulder joints.
Multi-structure bone segmentation in pediatric MR images with combined regularization from shape priors and adversarial network
Sep 15, 2020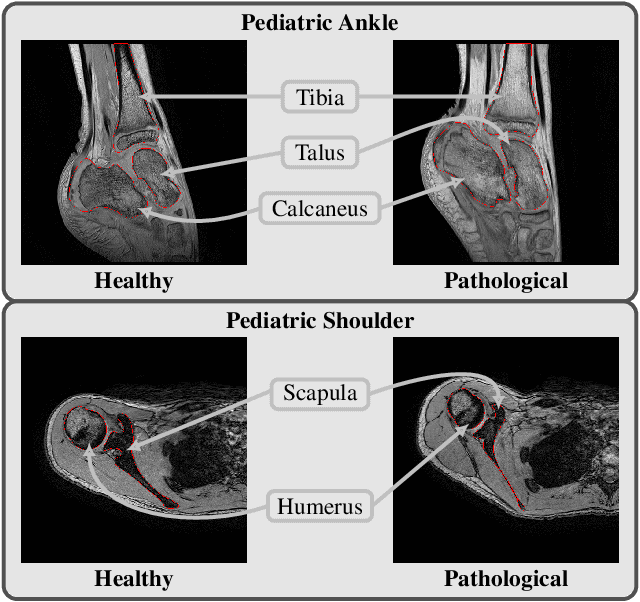

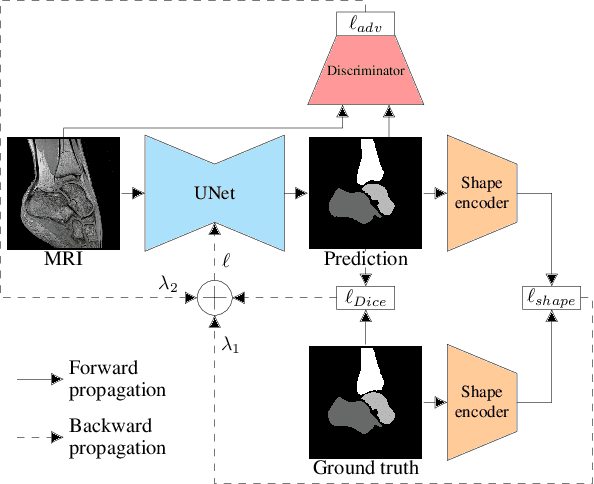

Abstract:Morphological and diagnostic evaluation of pediatric musculoskeletal system is crucial in clinical practice. However, most segmentation models do not perform well on scarce pediatric imaging data. We propose a regularized convolutional encoder-decoder network for the challenging task of segmenting pediatric magnetic resonance (MR) images. To overcome the scarcity and heterogeneity of pediatric imaging datasets, we adopt a regularization strategy to improve the generalization of segmentation models. To this end, we have conceived a novel optimization scheme for the segmentation network which comprises additional regularization terms to the loss function. In order to obtain globally consistent predictions, we incorporate a shape priors based regularization, derived from a non-linear shape representation learnt by an auto-encoder. Additionally, an adversarial regularization computed by a discriminator is integrated to encourage plausible delineations. Our method is evaluated for the task of multi-bone segmentation on two pediatric imaging datasets from different joints (ankle and shoulder), comprising pathological as well as healthy examinations. We illustrate that the proposed approach can be easily integrated into various multi-structure strategies and can improve the prediction accuracy of state-of-the-art models. The obtained results bring new perspectives for the management of pediatric musculoskeletal disorders.
Dynamic multi-object Gaussian process models: A framework for data-driven functional modelling of human joints
Jan 22, 2020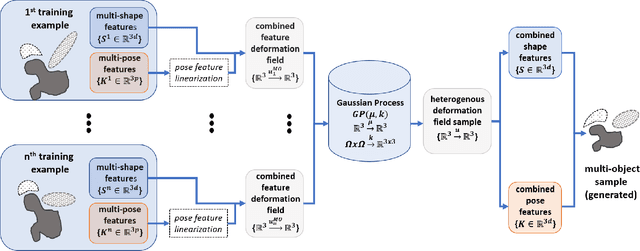
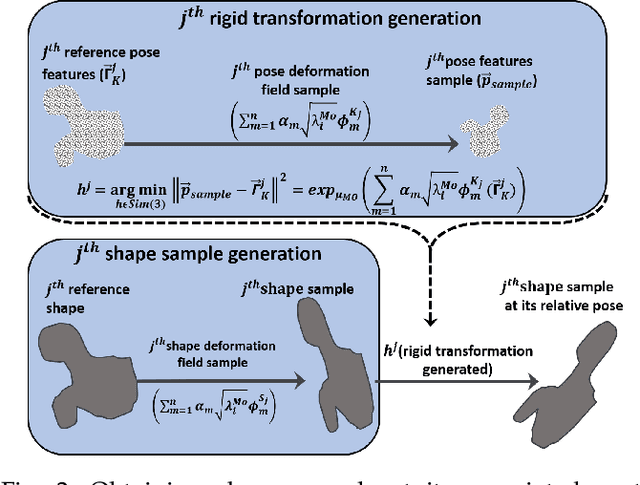
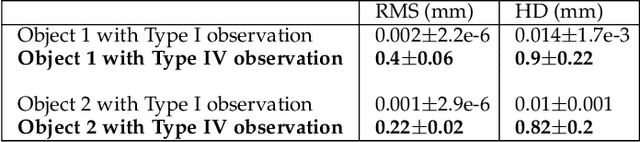
Abstract:Statistical shape models (SSMs) are state-of-the-art medical image analysis tools for extracting and explaining features across a set of biological structures. However, a principled and robust way to combine shape and pose features has been illusive due to three main issues: 1) Non-homogeneity of the data (data with linear and non-linear natural variation across features), 2) non-optimal representation of the $3D$ motion (rigid transformation representations that are not proportional to the kinetic energy that move an object from one position to the other), and 3) artificial discretization of the models. In this paper, we propose a new framework for dynamic multi-object statistical modelling framework for the analysis of human joints in a continuous domain. Specifically, we propose to normalise shape and dynamic spatial features in the same linearized statistical space permitting the use of linear statistics; we adopt an optimal 3D motion representation for more accurate rigid transformation comparisons; and we provide a 3D shape and pose prediction protocol using a Markov chain Monte Carlo sampling-based fitting. The framework affords an efficient generative dynamic multi-object modelling platform for biological joints. We validate the framework using a controlled synthetic data. Finally, the framework is applied to an analysis of the human shoulder joint to compare its performance with standard SSM approaches in prediction of shape while adding the advantage of determining relative pose between bones in a complex. Excellent validity is observed and the shoulder joint shape-pose prediction results suggest that the novel framework may have utility for a range of medical image analysis applications. Furthermore, the framework is generic and can be extended to n$>$2 objects, making it suitable for clinical and diagnostic methods for the management of joint disorders.
Combining Shape Priors with Conditional Adversarial Networks for Improved Scapula Segmentation in MR images
Oct 22, 2019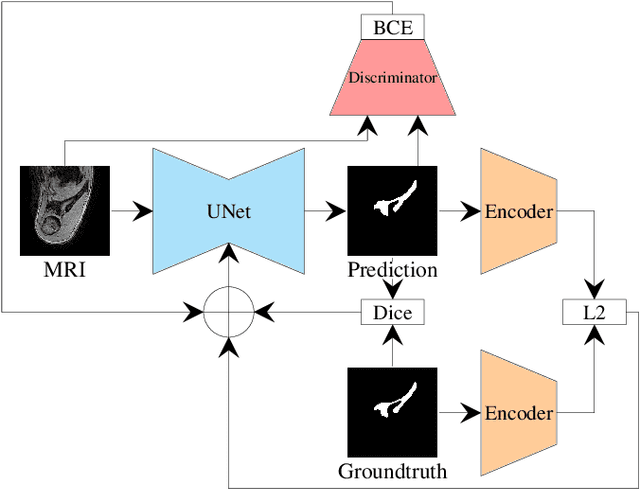
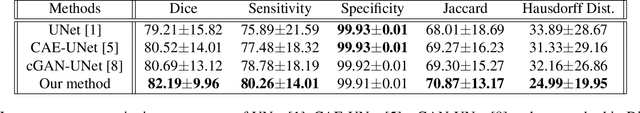
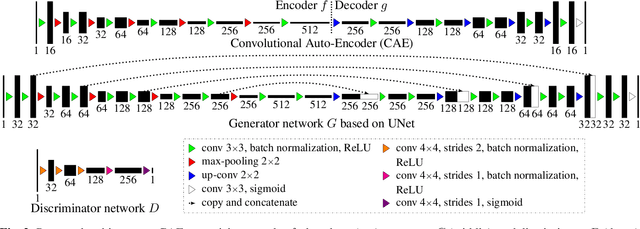
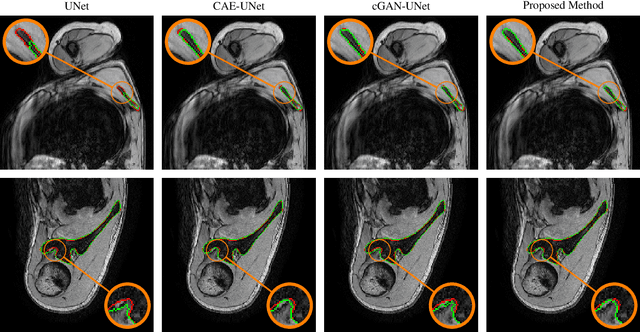
Abstract:This paper proposes an automatic method for scapula bone segmentation from Magnetic Resonance (MR) images using deep learning. The purpose of this work is to incorporate anatomical priors into a conditional adversarial framework, given a limited amount of heterogeneous annotated images. Our approach encourages the segmentation model to follow the global anatomical properties of the underlying anatomy through a learnt non-linear shape representation while the adversarial contribution refines the model by promoting realistic delineations. These contributions are evaluated on a data set of 15 pediatric shoulder examinations, and compared to state-of-the-art architectures including UNet and recent derivatives. The significant improvements achieved bring new perspectives for the pre-operative management of musculo-skeletal diseases.
Healthy versus pathological learning transferability in shoulder muscle MRI segmentation using deep convolutional encoder-decoders
Jan 06, 2019



Abstract:Automatic segmentation of pathological shoulder muscles in patients with musculo-skeletal diseases is a challenging task due to the huge variability in muscle shape, size, location, texture and injury. A reliable fully-automated segmentation method from magnetic resonance images could greatly help clinicians to plan therapeutic interventions and predict interventional outcomes while eliminating time consuming manual segmentation efforts. The purpose of this work is three-fold. First, we investigate the feasibility of pathological shoulder muscle segmentation using deep learning techniques, given a very limited amount of available annotated pediatric data. Second, we address the learning transferability from healthy to pathological data by comparing different learning schemes in terms of model generalizability. Third, extended versions of deep convolutional encoder-decoder architectures using encoders pre-trained on non-medical data are proposed to improve the segmentation accuracy. Methodological aspects are evaluated in a leave-one-out fashion on a dataset of 24 shoulder examinations from patients with obstetrical brachial plexus palsy and focus on 4 different muscles including deltoid as well as infraspinatus, supraspinatus and subscapularis from the rotator cuff. The most relevant segmentation model is partially pre-trained on ImageNet and jointly exploits inter-patient healthy and pathological annotated data. Its performance reaches Dice scores of 82.4%, 82.0%, 71.0% and 82.8% for deltoid, infraspinatus, supraspinatus and subscapularis muscles. Absolute surface estimation errors are all below 83mm$^2$ except for supraspinatus with 134.6mm$^2$. These contributions offer new perspectives for force inference in the context of musculo-skeletal disorder management.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge