Christelle Pons
Generalizable multi-task, multi-domain deep segmentation of sparse pediatric imaging datasets via multi-scale contrastive regularization and multi-joint anatomical priors
Jul 27, 2022


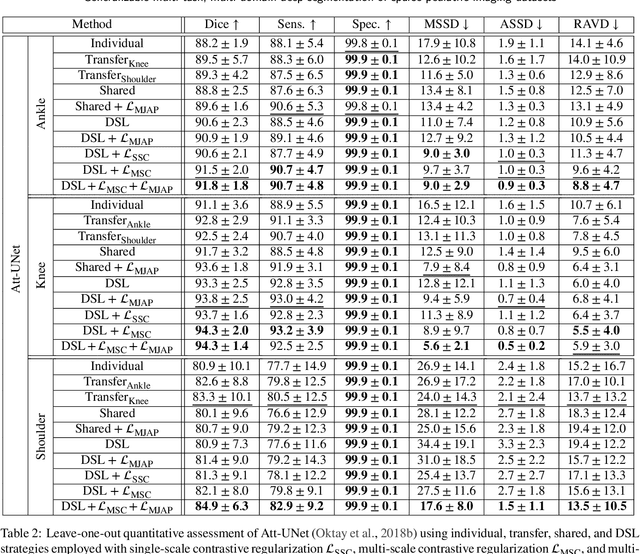
Abstract:Clinical diagnosis of the pediatric musculoskeletal system relies on the analysis of medical imaging examinations. In the medical image processing pipeline, semantic segmentation using deep learning algorithms enables an automatic generation of patient-specific three-dimensional anatomical models which are crucial for morphological evaluation. However, the scarcity of pediatric imaging resources may result in reduced accuracy and generalization performance of individual deep segmentation models. In this study, we propose to design a novel multi-task, multi-domain learning framework in which a single segmentation network is optimized over the union of multiple datasets arising from distinct parts of the anatomy. Unlike previous approaches, we simultaneously consider multiple intensity domains and segmentation tasks to overcome the inherent scarcity of pediatric data while leveraging shared features between imaging datasets. To further improve generalization capabilities, we employ a transfer learning scheme from natural image classification, along with a multi-scale contrastive regularization aimed at promoting domain-specific clusters in the shared representations, and multi-joint anatomical priors to enforce anatomically consistent predictions. We evaluate our contributions for performing bone segmentation using three scarce and pediatric imaging datasets of the ankle, knee, and shoulder joints. Our results demonstrate that the proposed approach outperforms individual, transfer, and shared segmentation schemes in Dice metric with statistically sufficient margins. The proposed model brings new perspectives towards intelligent use of imaging resources and better management of pediatric musculoskeletal disorders.
Multi-Task, Multi-Domain Deep Segmentation with Shared Representations and Contrastive Regularization for Sparse Pediatric Datasets
May 21, 2021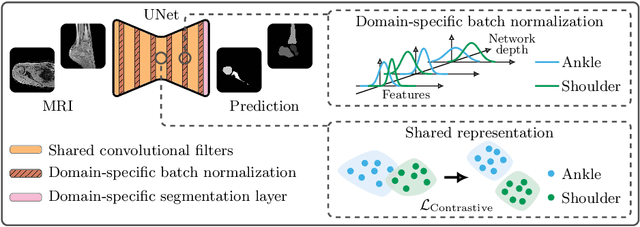



Abstract:Automatic segmentation of magnetic resonance (MR) images is crucial for morphological evaluation of the pediatric musculoskeletal system in clinical practice. However, the accuracy and generalization performance of individual segmentation models are limited due to the restricted amount of annotated pediatric data. Hence, we propose to train a segmentation model on multiple datasets, arising from different parts of the anatomy, in a multi-task and multi-domain learning framework. This approach allows to overcome the inherent scarcity of pediatric data while benefiting from a more robust shared representation. The proposed segmentation network comprises shared convolutional filters, domain-specific batch normalization parameters that compute the respective dataset statistics and a domain-specific segmentation layer. Furthermore, a supervised contrastive regularization is integrated to further improve generalization capabilities, by promoting intra-domain similarity and impose inter-domain margins in embedded space. We evaluate our contributions on two pediatric imaging datasets of the ankle and shoulder joints for bone segmentation. Results demonstrate that the proposed model outperforms state-of-the-art approaches.
Multi-Structure Deep Segmentation with Shape Priors and Latent Adversarial Regularization
Jan 25, 2021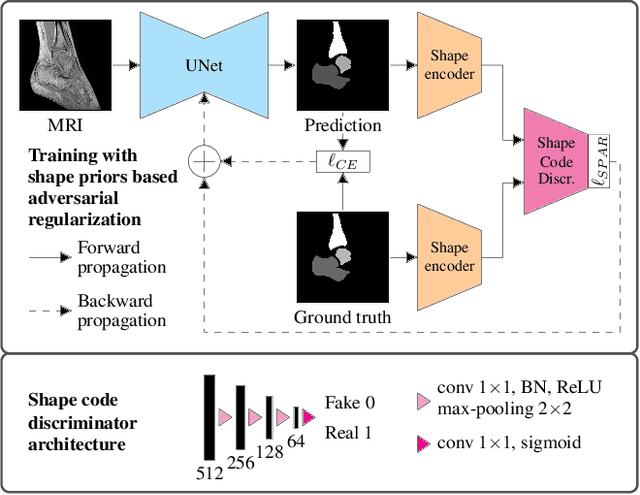


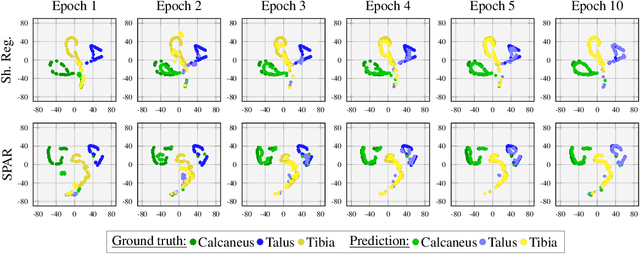
Abstract:Automatic segmentation of the musculoskeletal system in pediatric magnetic resonance (MR) images is a challenging but crucial task for morphological evaluation in clinical practice. We propose a deep learning-based regularized segmentation method for multi-structure bone delineation in MR images, designed to overcome the inherent scarcity and heterogeneity of pediatric data. Based on a newly devised shape code discriminator, our adversarial regularization scheme enforces the deep network to follow a learnt shape representation of the anatomy. The novel shape priors based adversarial regularization (SPAR) exploits latent shape codes arising from ground truth and predicted masks to guide the segmentation network towards more consistent and plausible predictions. Our contribution is compared to state-of-the-art regularization methods on two pediatric musculoskeletal imaging datasets from ankle and shoulder joints.
Healthy versus pathological learning transferability in shoulder muscle MRI segmentation using deep convolutional encoder-decoders
Jan 06, 2019



Abstract:Automatic segmentation of pathological shoulder muscles in patients with musculo-skeletal diseases is a challenging task due to the huge variability in muscle shape, size, location, texture and injury. A reliable fully-automated segmentation method from magnetic resonance images could greatly help clinicians to plan therapeutic interventions and predict interventional outcomes while eliminating time consuming manual segmentation efforts. The purpose of this work is three-fold. First, we investigate the feasibility of pathological shoulder muscle segmentation using deep learning techniques, given a very limited amount of available annotated pediatric data. Second, we address the learning transferability from healthy to pathological data by comparing different learning schemes in terms of model generalizability. Third, extended versions of deep convolutional encoder-decoder architectures using encoders pre-trained on non-medical data are proposed to improve the segmentation accuracy. Methodological aspects are evaluated in a leave-one-out fashion on a dataset of 24 shoulder examinations from patients with obstetrical brachial plexus palsy and focus on 4 different muscles including deltoid as well as infraspinatus, supraspinatus and subscapularis from the rotator cuff. The most relevant segmentation model is partially pre-trained on ImageNet and jointly exploits inter-patient healthy and pathological annotated data. Its performance reaches Dice scores of 82.4%, 82.0%, 71.0% and 82.8% for deltoid, infraspinatus, supraspinatus and subscapularis muscles. Absolute surface estimation errors are all below 83mm$^2$ except for supraspinatus with 134.6mm$^2$. These contributions offer new perspectives for force inference in the context of musculo-skeletal disorder management.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge