Tinashe E. M. Mutsvangwa
Bridging the Gap: Generalising State-of-the-Art U-Net Models to Sub-Saharan African Populations
Dec 19, 2023Abstract:A critical challenge for tumour segmentation models is the ability to adapt to diverse clinical settings, particularly when applied to poor-quality neuroimaging data. The uncertainty surrounding this adaptation stems from the lack of representative datasets, leaving top-performing models without exposure to common artifacts found in MRI data throughout Sub-Saharan Africa (SSA). We replicated a framework that secured the 2nd position in the 2022 BraTS competition to investigate the impact of dataset composition on model performance and pursued four distinct approaches through training a model with: 1) BraTS-Africa data only (train_SSA, N=60), 2) BraTS-Adult Glioma data only (train_GLI, N=1251), 3) both datasets together (train_ALL, N=1311), and 4) through further training the train_GLI model with BraTS-Africa data (train_ftSSA). Notably, training on a smaller low-quality dataset alone (train_SSA) yielded subpar results, and training on a larger high-quality dataset alone (train_GLI) struggled to delineate oedematous tissue in the low-quality validation set. The most promising approach (train_ftSSA) involved pre-training a model on high-quality neuroimages and then fine-tuning it on the smaller, low-quality dataset. This approach outperformed the others, ranking second in the MICCAI BraTS Africa global challenge external testing phase. These findings underscore the significance of larger sample sizes and broad exposure to data in improving segmentation performance. Furthermore, we demonstrated that there is potential for improving such models by fine-tuning them with a wider range of data locally.
Dynamic multi feature-class Gaussian process models
Dec 08, 2021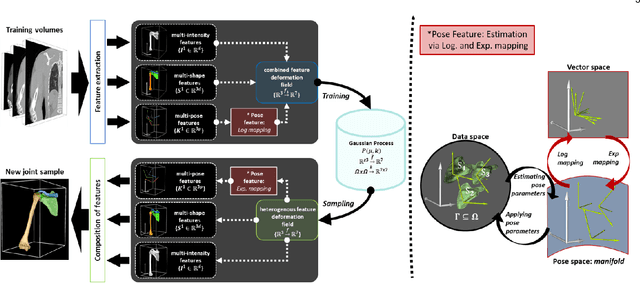
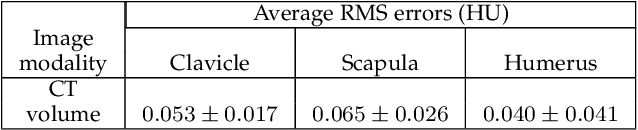
Abstract:In model-based medical image analysis, three features of interest are the shape of structures of interest, their relative pose, and image intensity profiles representative of some physical property. Often, these are modelled separately through statistical models by decomposing the object's features into a set of basis functions through principal geodesic analysis or principal component analysis. This study presents a statistical modelling method for automatic learning of shape, pose and intensity features in medical images which we call the Dynamic multi feature-class Gaussian process models (DMFC-GPM). A DMFC-GPM is a Gaussian process (GP)-based model with a shared latent space that encodes linear and non-linear variation. Our method is defined in a continuous domain with a principled way to represent shape, pose and intensity feature classes in a linear space, based on deformation fields. A deformation field-based metric is adapted in the method for modelling shape and intensity feature variation as well as for comparing rigid transformations (pose). Moreover, DMFC-GPMs inherit properties intrinsic to GPs including marginalisation and regression. Furthermore, they allow for adding additional pose feature variability on top of those obtained from the image acquisition process; what we term as permutation modelling. For image analysis tasks using DMFC-GPMs, we adapt Metropolis-Hastings algorithms making the prediction of features fully probabilistic. We validate the method using controlled synthetic data and we perform experiments on bone structures from CT images of the shoulder to illustrate the efficacy of the model for pose and shape feature prediction. The model performance results suggest that this new modelling paradigm is robust, accurate, accessible, and has potential applications including the management of musculoskeletal disorders and clinical decision making
Dynamic multi-object Gaussian process models: A framework for data-driven functional modelling of human joints
Jan 22, 2020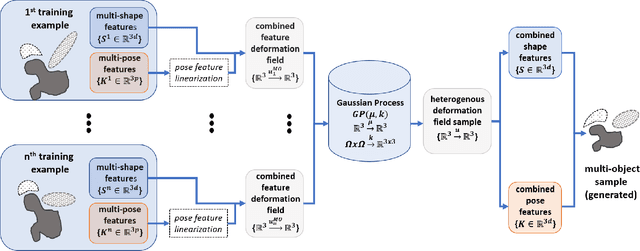
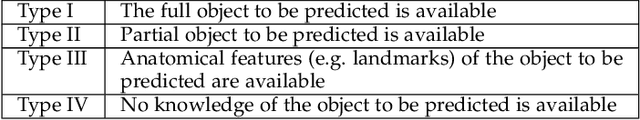
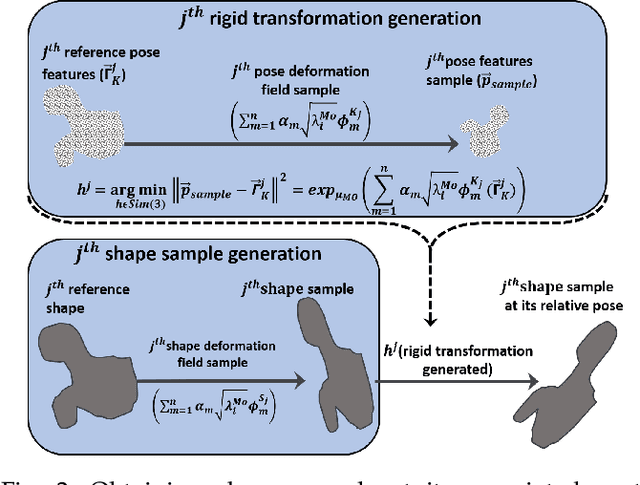
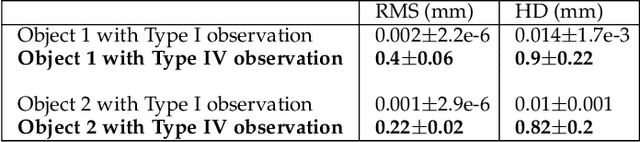
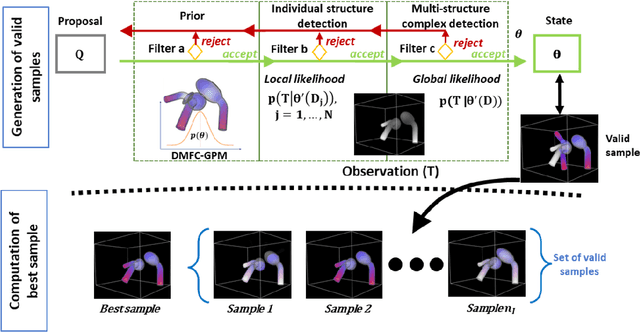
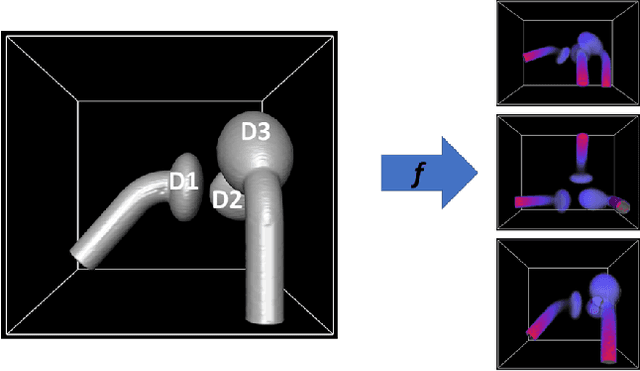
Abstract:Statistical shape models (SSMs) are state-of-the-art medical image analysis tools for extracting and explaining features across a set of biological structures. However, a principled and robust way to combine shape and pose features has been illusive due to three main issues: 1) Non-homogeneity of the data (data with linear and non-linear natural variation across features), 2) non-optimal representation of the $3D$ motion (rigid transformation representations that are not proportional to the kinetic energy that move an object from one position to the other), and 3) artificial discretization of the models. In this paper, we propose a new framework for dynamic multi-object statistical modelling framework for the analysis of human joints in a continuous domain. Specifically, we propose to normalise shape and dynamic spatial features in the same linearized statistical space permitting the use of linear statistics; we adopt an optimal 3D motion representation for more accurate rigid transformation comparisons; and we provide a 3D shape and pose prediction protocol using a Markov chain Monte Carlo sampling-based fitting. The framework affords an efficient generative dynamic multi-object modelling platform for biological joints. We validate the framework using a controlled synthetic data. Finally, the framework is applied to an analysis of the human shoulder joint to compare its performance with standard SSM approaches in prediction of shape while adding the advantage of determining relative pose between bones in a complex. Excellent validity is observed and the shoulder joint shape-pose prediction results suggest that the novel framework may have utility for a range of medical image analysis applications. Furthermore, the framework is generic and can be extended to n$>$2 objects, making it suitable for clinical and diagnostic methods for the management of joint disorders.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge