Udunna C. Anazodo
Empowering Medical Equipment Sustainability in Low-Resource Settings: An AI-Powered Diagnostic and Support Platform for Biomedical Technicians
Jan 23, 2026Abstract:In low- and middle-income countries (LMICs), a significant proportion of medical diagnostic equipment remains underutilized or non-functional due to a lack of timely maintenance, limited access to technical expertise, and minimal support from manufacturers, particularly for devices acquired through third-party vendors or donations. This challenge contributes to increased equipment downtime, delayed diagnoses, and compromised patient care. This research explores the development and validation of an AI-powered support platform designed to assist biomedical technicians in diagnosing and repairing medical devices in real-time. The system integrates a large language model (LLM) with a user-friendly web interface, enabling imaging technologists/radiographers and biomedical technicians to input error codes or device symptoms and receive accurate, step-by-step troubleshooting guidance. The platform also includes a global peer-to-peer discussion forum to support knowledge exchange and provide additional context for rare or undocumented issues. A proof of concept was developed using the Philips HDI 5000 ultrasound machine, achieving 100% precision in error code interpretation and 80% accuracy in suggesting corrective actions. This study demonstrates the feasibility and potential of AI-driven systems to support medical device maintenance, with the aim of reducing equipment downtime to improve healthcare delivery in resource-constrained environments.
Towards Trustworthy Breast Tumor Segmentation in Ultrasound using Monte Carlo Dropout and Deep Ensembles for Epistemic Uncertainty Estimation
Aug 25, 2025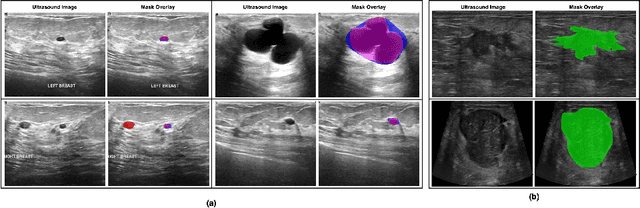
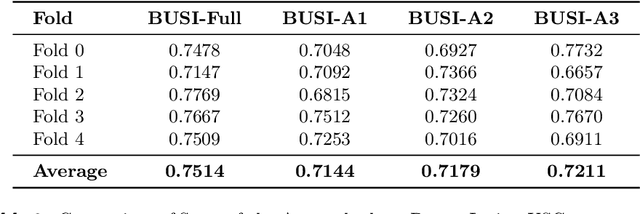
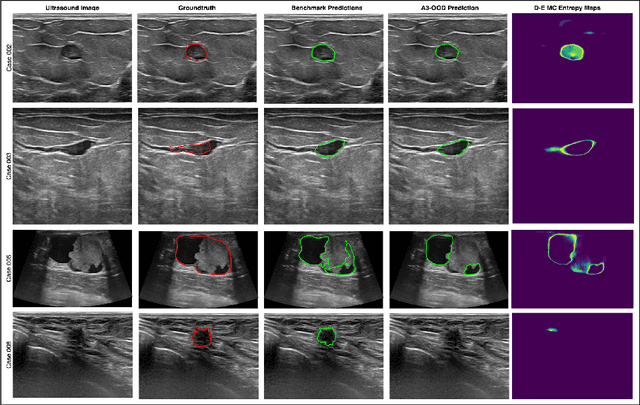

Abstract:Automated segmentation of BUS images is important for precise lesion delineation and tumor characterization, but is challenged by inherent artifacts and dataset inconsistencies. In this work, we evaluate the use of a modified Residual Encoder U-Net for breast ultrasound segmentation, with a focus on uncertainty quantification. We identify and correct for data duplication in the BUSI dataset, and use a deduplicated subset for more reliable estimates of generalization performance. Epistemic uncertainty is quantified using Monte Carlo dropout, deep ensembles, and their combination. Models are benchmarked on both in-distribution and out-of-distribution datasets to demonstrate how they generalize to unseen cross-domain data. Our approach achieves state-of-the-art segmentation accuracy on the Breast-Lesion-USG dataset with in-distribution validation, and provides calibrated uncertainty estimates that effectively signal regions of low model confidence. Performance declines and increased uncertainty observed in out-of-distribution evaluation highlight the persistent challenge of domain shift in medical imaging, and the importance of integrated uncertainty modeling for trustworthy clinical deployment. \footnote{Code available at: https://github.com/toufiqmusah/nn-uncertainty.git}
Generative Style Transfer for MRI Image Segmentation: A Case of Glioma Segmentation in Sub-Saharan Africa
Jan 07, 2025Abstract:In Sub-Saharan Africa (SSA), the utilization of lower-quality Magnetic Resonance Imaging (MRI) technology raises questions about the applicability of machine learning methods for clinical tasks. This study aims to provide a robust deep learning-based brain tumor segmentation (BraTS) method tailored for the SSA population using a threefold approach. Firstly, the impact of domain shift from the SSA training data on model efficacy was examined, revealing no significant effect. Secondly, a comparative analysis of 3D and 2D full-resolution models using the nnU-Net framework indicates similar performance of both the models trained for 300 epochs achieving a five-fold cross-validation score of 0.93. Lastly, addressing the performance gap observed in SSA validation as opposed to the relatively larger BraTS glioma (GLI) validation set, two strategies are proposed: fine-tuning SSA cases using the GLI+SSA best-pretrained 2D fullres model at 300 epochs, and introducing a novel neural style transfer-based data augmentation technique for the SSA cases. This investigation underscores the potential of enhancing brain tumor prediction within SSA's unique healthcare landscape.
Pseudo-MRI-Guided PET Image Reconstruction Method Based on a Diffusion Probabilistic Model
Mar 26, 2024



Abstract:Anatomically guided PET reconstruction using MRI information has been shown to have the potential to improve PET image quality. However, these improvements are limited to PET scans with paired MRI information. In this work we employed a diffusion probabilistic model (DPM) to infer T1-weighted-MRI (deep-MRI) images from FDG-PET brain images. We then use the DPM-generated T1w-MRI to guide the PET reconstruction. The model was trained with brain FDG scans, and tested in datasets containing multiple levels of counts. Deep-MRI images appeared somewhat degraded than the acquired MRI images. Regarding PET image quality, volume of interest analysis in different brain regions showed that both PET reconstructed images using the acquired and the deep-MRI images improved image quality compared to OSEM. Same conclusions were found analysing the decimated datasets. A subjective evaluation performed by two physicians confirmed that OSEM scored consistently worse than the MRI-guided PET images and no significant differences were observed between the MRI-guided PET images. This proof of concept shows that it is possible to infer DPM-based MRI imagery to guide the PET reconstruction, enabling the possibility of changing reconstruction parameters such as the strength of the prior on anatomically guided PET reconstruction in the absence of MRI.
Bridging the Gap: Generalising State-of-the-Art U-Net Models to Sub-Saharan African Populations
Dec 19, 2023Abstract:A critical challenge for tumour segmentation models is the ability to adapt to diverse clinical settings, particularly when applied to poor-quality neuroimaging data. The uncertainty surrounding this adaptation stems from the lack of representative datasets, leaving top-performing models without exposure to common artifacts found in MRI data throughout Sub-Saharan Africa (SSA). We replicated a framework that secured the 2nd position in the 2022 BraTS competition to investigate the impact of dataset composition on model performance and pursued four distinct approaches through training a model with: 1) BraTS-Africa data only (train_SSA, N=60), 2) BraTS-Adult Glioma data only (train_GLI, N=1251), 3) both datasets together (train_ALL, N=1311), and 4) through further training the train_GLI model with BraTS-Africa data (train_ftSSA). Notably, training on a smaller low-quality dataset alone (train_SSA) yielded subpar results, and training on a larger high-quality dataset alone (train_GLI) struggled to delineate oedematous tissue in the low-quality validation set. The most promising approach (train_ftSSA) involved pre-training a model on high-quality neuroimages and then fine-tuning it on the smaller, low-quality dataset. This approach outperformed the others, ranking second in the MICCAI BraTS Africa global challenge external testing phase. These findings underscore the significance of larger sample sizes and broad exposure to data in improving segmentation performance. Furthermore, we demonstrated that there is potential for improving such models by fine-tuning them with a wider range of data locally.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge