Raymond Confidence
SharpXR: Structure-Aware Denoising for Pediatric Chest X-Rays
Aug 11, 2025Abstract:Pediatric chest X-ray imaging is essential for early diagnosis, particularly in low-resource settings where advanced imaging modalities are often inaccessible. Low-dose protocols reduce radiation exposure in children but introduce substantial noise that can obscure critical anatomical details. Conventional denoising methods often degrade fine details, compromising diagnostic accuracy. In this paper, we present SharpXR, a structure-aware dual-decoder U-Net designed to denoise low-dose pediatric X-rays while preserving diagnostically relevant features. SharpXR combines a Laplacian-guided edge-preserving decoder with a learnable fusion module that adaptively balances noise suppression and structural detail retention. To address the scarcity of paired training data, we simulate realistic Poisson-Gaussian noise on the Pediatric Pneumonia Chest X-ray dataset. SharpXR outperforms state-of-the-art baselines across all evaluation metrics while maintaining computational efficiency suitable for resource-constrained settings. SharpXR-denoised images improved downstream pneumonia classification accuracy from 88.8% to 92.5%, underscoring its diagnostic value in low-resource pediatric care.
Deep Ensemble approach for Enhancing Brain Tumor Segmentation in Resource-Limited Settings
Feb 04, 2025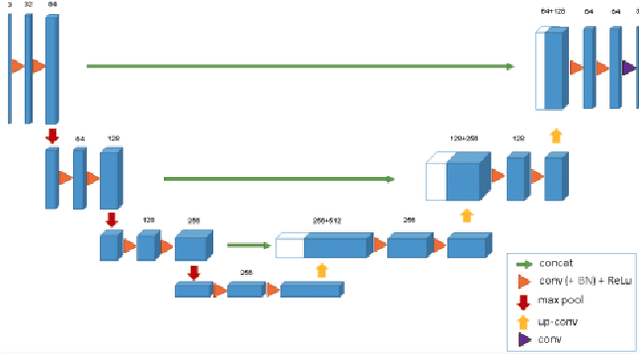
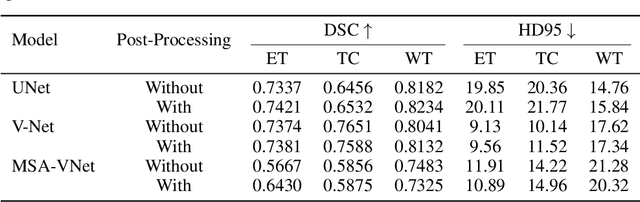
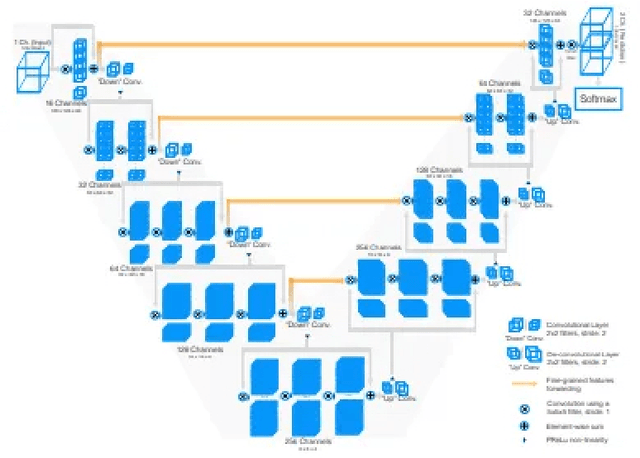
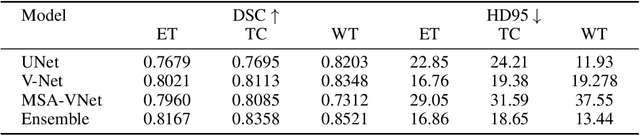
Abstract:Segmentation of brain tumors is a critical step in treatment planning, yet manual segmentation is both time-consuming and subjective, relying heavily on the expertise of radiologists. In Sub-Saharan Africa, this challenge is magnified by overburdened medical systems and limited access to advanced imaging modalities and expert radiologists. Automating brain tumor segmentation using deep learning offers a promising solution. Convolutional Neural Networks (CNNs), especially the U-Net architecture, have shown significant potential. However, a major challenge remains: achieving generalizability across different datasets. This study addresses this gap by developing a deep learning ensemble that integrates UNet3D, V-Net, and MSA-VNet models for the semantic segmentation of gliomas. By initially training on the BraTS-GLI dataset and fine-tuning with the BraTS-SSA dataset, we enhance model performance. Our ensemble approach significantly outperforms individual models, achieving DICE scores of 0.8358 for Tumor Core, 0.8521 for Whole Tumor, and 0.8167 for Enhancing Tumor. These results underscore the potential of ensemble methods in improving the accuracy and reliability of automated brain tumor segmentation, particularly in resource-limited settings.
Generative Style Transfer for MRI Image Segmentation: A Case of Glioma Segmentation in Sub-Saharan Africa
Jan 07, 2025Abstract:In Sub-Saharan Africa (SSA), the utilization of lower-quality Magnetic Resonance Imaging (MRI) technology raises questions about the applicability of machine learning methods for clinical tasks. This study aims to provide a robust deep learning-based brain tumor segmentation (BraTS) method tailored for the SSA population using a threefold approach. Firstly, the impact of domain shift from the SSA training data on model efficacy was examined, revealing no significant effect. Secondly, a comparative analysis of 3D and 2D full-resolution models using the nnU-Net framework indicates similar performance of both the models trained for 300 epochs achieving a five-fold cross-validation score of 0.93. Lastly, addressing the performance gap observed in SSA validation as opposed to the relatively larger BraTS glioma (GLI) validation set, two strategies are proposed: fine-tuning SSA cases using the GLI+SSA best-pretrained 2D fullres model at 300 epochs, and introducing a novel neural style transfer-based data augmentation technique for the SSA cases. This investigation underscores the potential of enhancing brain tumor prediction within SSA's unique healthcare landscape.
Towards SAMBA: Segment Anything Model for Brain Tumor Segmentation in Sub-Sharan African Populations
Dec 19, 2023



Abstract:Gliomas, the most prevalent primary brain tumors, require precise segmentation for diagnosis and treatment planning. However, this task poses significant challenges, particularly in the African population, were limited access to high-quality imaging data hampers algorithm performance. In this study, we propose an innovative approach combining the Segment Anything Model (SAM) and a voting network for multi-modal glioma segmentation. By fine-tuning SAM with bounding box-guided prompts (SAMBA), we adapt the model to the complexities of African datasets. Our ensemble strategy, utilizing multiple modalities and views, produces a robust consensus segmentation, addressing intra-tumoral heterogeneity. Although the low quality of scans presents difficulties, our methodology has the potential to profoundly impact clinical practice in resource-limited settings such as Africa, improving treatment decisions and advancing neuro-oncology research. Furthermore, successful application to other brain tumor types and lesions in the future holds promise for a broader transformation in neurological imaging, improving healthcare outcomes across all settings. This study was conducted on the Brain Tumor Segmentation (BraTS) Challenge Africa (BraTS-Africa) dataset, which provides a valuable resource for addressing challenges specific to resource-limited settings, particularly the African population, and facilitating the development of effective and more generalizable segmentation algorithms. To illustrate our approach's potential, our experiments on the BraTS-Africa dataset yielded compelling results, with SAM attaining a Dice coefficient of 86.6 for binary segmentation and 60.4 for multi-class segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge