Eric Stindel
Laboratory of Medical Information Processing
Fully automated workflow for the design of patient-specific orthopaedic implants: application to total knee arthroplasty
Mar 25, 2024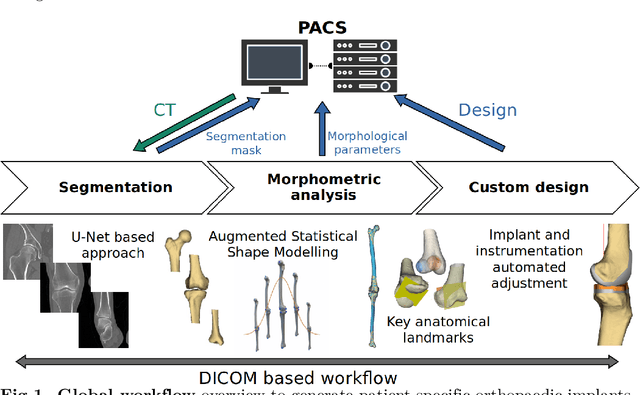
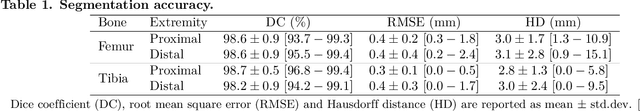
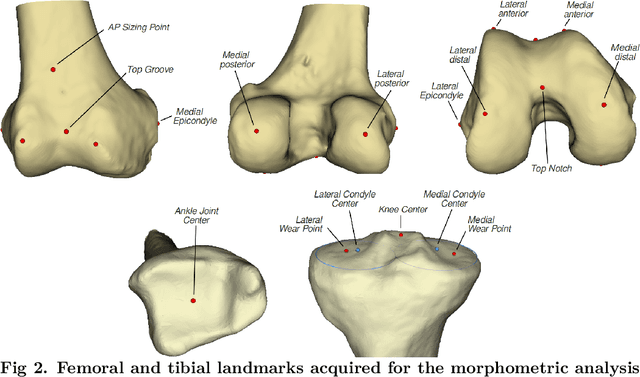
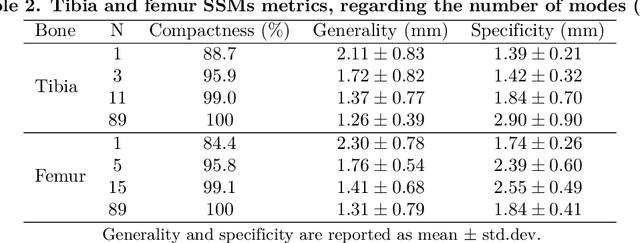
Abstract:Arthroplasty is commonly performed to treat joint osteoarthritis, reducing pain and improving mobility. While arthroplasty has known several technical improvements, a significant share of patients are still unsatisfied with their surgery. Personalised arthroplasty improves surgical outcomes however current solutions require delays, making it difficult to integrate in clinical routine. We propose a fully automated workflow to design patient-specific implants, presented for total knee arthroplasty, the most widely performed arthroplasty in the world nowadays. The proposed pipeline first uses artificial neural networks to segment the proximal and distal extremities of the femur and tibia. Then the full bones are reconstructed using augmented statistical shape models, combining shape and landmarks information. Finally, 77 morphological parameters are computed to design patient-specific implants. The developed workflow has been trained using 91 CT scans of lower limb and evaluated on 41 CT scans manually segmented, in terms of accuracy and execution time. The workflow accuracy was $0.4\pm0.2mm$ for the segmentation, $1.2\pm0.4mm$ for the full bones reconstruction, and $2.8\pm2.2mm$ for the anatomical landmarks determination. The custom implants fitted the patients' anatomy with $0.6\pm0.2mm$ accuracy. The whole process from segmentation to implants' design lasted about 5 minutes. The proposed workflow allows for a fast and reliable personalisation of knee implants, directly from the patient CT image without requiring any manual intervention. It establishes a patient-specific pre-operative planning for TKA in a very short time making it easily available for all patients. Combined with efficient implant manufacturing techniques, this solution could help answer the growing number of arthroplasties while reducing complications and improving the patients' satisfaction.
Making the best of data derived from a daily practice in clinical legal medicine for research and practice - the example of Spe3dLab
Jul 26, 2017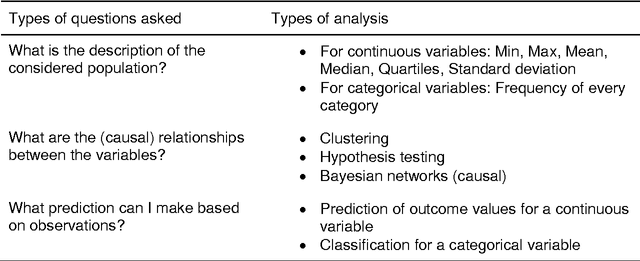
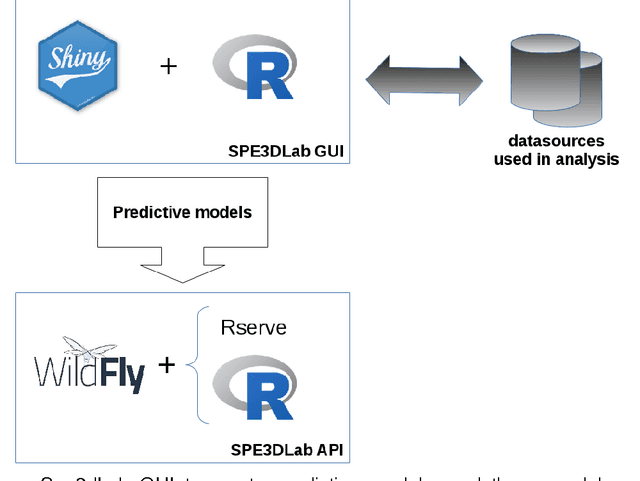
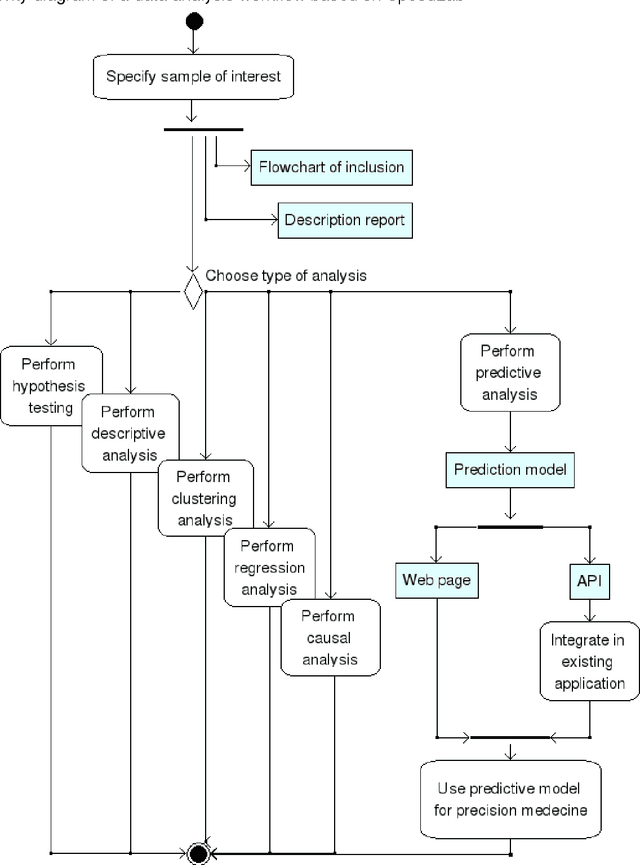
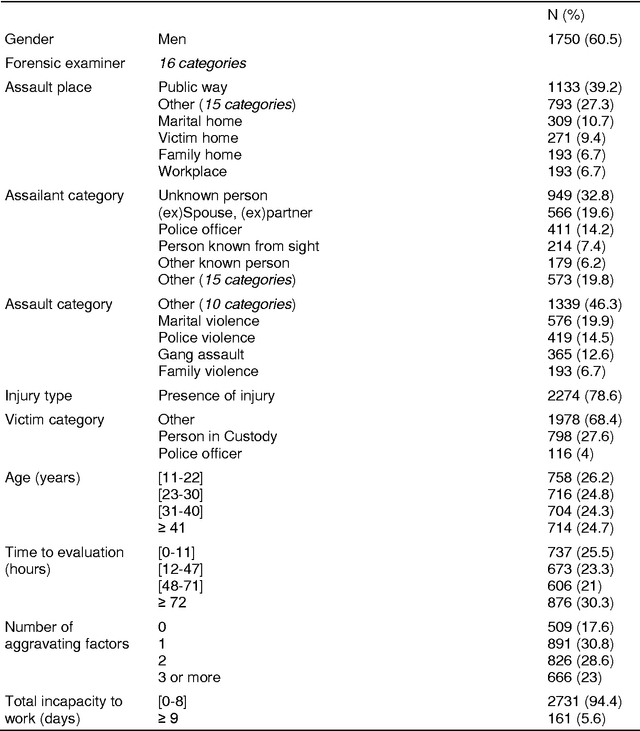
Abstract:Forensic science suffers from a lack of studies with high-quality design, such as randomized controlled trials (RCT). Evidence in forensic science may be of insufficient quality, which is a major concern. Results from RCT are criticized for providing artificial results that are not useful in real life and unfit for individualized prescription. Various sources of collected data (e.g. data collected in routine practice) could be exploited for distinct goals. Obstacles remain before such data can be practically accessed and used, including technical issues. We present an easy-to-use software dedicated to innovative data analyses for practitioners and researchers. We provide 2 examples in forensics. Spe3dLab has been developed by 3 French teams: a bioinformatics laboratory (LaTIM), a private partner (Tekliko) and a department of forensic medicine (Jean Verdier Hospital). It was designed to be open source, relying on documented and maintained libraries, query-oriented and capable of handling the entire data process from capture to export of best predictive models for their integration in information systems. Spe3dLab was used for 2 specific forensics applications: i) the search for multiple causal factors and ii) the best predictive model of the functional impairment (total incapacity to work, TIW) of assault survivors. 2,892 patients were included over a 6-month period. Time to evaluation was the only direct cause identified for TIW, and victim category was an indirect cause. The specificity and sensitivity of the predictive model were 99.9% and 90%, respectively. Spe3dLab is a quick and efficient tool for accessing observational, routinely collected data and performing innovative analyses. Analyses can be exported for validation and routine use by practitioners, e.g., for computer-aided evaluation of complex problems. It can provide a fully integrated solution for individualized medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge