Thomas Savage
Self-Reported Confidence of Large Language Models in Gastroenterology: Analysis of Commercial, Open-Source, and Quantized Models
Mar 24, 2025Abstract:This study evaluated self-reported response certainty across several large language models (GPT, Claude, Llama, Phi, Mistral, Gemini, Gemma, and Qwen) using 300 gastroenterology board-style questions. The highest-performing models (GPT-o1 preview, GPT-4o, and Claude-3.5-Sonnet) achieved Brier scores of 0.15-0.2 and AUROC of 0.6. Although newer models demonstrated improved performance, all exhibited a consistent tendency towards overconfidence. Uncertainty estimation presents a significant challenge to the safe use of LLMs in healthcare. Keywords: Large Language Models; Confidence Elicitation; Artificial Intelligence; Gastroenterology; Uncertainty Quantification
Large Language Models versus Classical Machine Learning: Performance in COVID-19 Mortality Prediction Using High-Dimensional Tabular Data
Sep 02, 2024



Abstract:Background: This study aimed to evaluate and compare the performance of classical machine learning models (CMLs) and large language models (LLMs) in predicting mortality associated with COVID-19 by utilizing a high-dimensional tabular dataset. Materials and Methods: We analyzed data from 9,134 COVID-19 patients collected across four hospitals. Seven CML models, including XGBoost and random forest (RF), were trained and evaluated. The structured data was converted into text for zero-shot classification by eight LLMs, including GPT-4 and Mistral-7b. Additionally, Mistral-7b was fine-tuned using the QLoRA approach to enhance its predictive capabilities. Results: Among the CML models, XGBoost and RF achieved the highest accuracy, with F1 scores of 0.87 for internal validation and 0.83 for external validation. In the LLM category, GPT-4 was the top performer with an F1 score of 0.43. Fine-tuning Mistral-7b significantly improved its recall from 1% to 79%, resulting in an F1 score of 0.74, which was stable during external validation. Conclusion: While LLMs show moderate performance in zero-shot classification, fine-tuning can significantly enhance their effectiveness, potentially aligning them closer to CML models. However, CMLs still outperform LLMs in high-dimensional tabular data tasks.
Methods to Estimate Large Language Model Confidence
Dec 08, 2023



Abstract:Large Language Models have difficulty communicating uncertainty, which is a significant obstacle to applying LLMs to complex medical tasks. This study evaluates methods to measure LLM confidence when suggesting a diagnosis for challenging clinical vignettes. GPT4 was asked a series of challenging case questions using Chain of Thought and Self Consistency prompting. Multiple methods were investigated to assess model confidence and evaluated on their ability to predict the models observed accuracy. The methods evaluated were Intrinsic Confidence, SC Agreement Frequency and CoT Response Length. SC Agreement Frequency correlated with observed accuracy, yielding a higher Area under the Receiver Operating Characteristic Curve compared to Intrinsic Confidence and CoT Length analysis. SC agreement is the most useful proxy for model confidence, especially for medical diagnosis. Model Intrinsic Confidence and CoT Response Length exhibit a weaker ability to differentiate between correct and incorrect answers, preventing them from being reliable and interpretable markers for model confidence. We conclude GPT4 has a limited ability to assess its own diagnostic accuracy. SC Agreement Frequency is the most useful method to measure GPT4 confidence.
MedAlign: A Clinician-Generated Dataset for Instruction Following with Electronic Medical Records
Aug 27, 2023
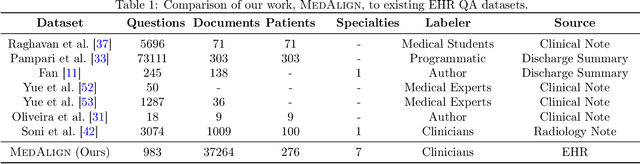
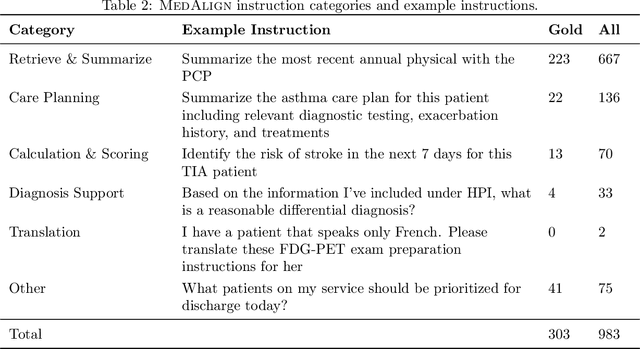

Abstract:The ability of large language models (LLMs) to follow natural language instructions with human-level fluency suggests many opportunities in healthcare to reduce administrative burden and improve quality of care. However, evaluating LLMs on realistic text generation tasks for healthcare remains challenging. Existing question answering datasets for electronic health record (EHR) data fail to capture the complexity of information needs and documentation burdens experienced by clinicians. To address these challenges, we introduce MedAlign, a benchmark dataset of 983 natural language instructions for EHR data. MedAlign is curated by 15 clinicians (7 specialities), includes clinician-written reference responses for 303 instructions, and provides 276 longitudinal EHRs for grounding instruction-response pairs. We used MedAlign to evaluate 6 general domain LLMs, having clinicians rank the accuracy and quality of each LLM response. We found high error rates, ranging from 35% (GPT-4) to 68% (MPT-7B-Instruct), and an 8.3% drop in accuracy moving from 32k to 2k context lengths for GPT-4. Finally, we report correlations between clinician rankings and automated natural language generation metrics as a way to rank LLMs without human review. We make MedAlign available under a research data use agreement to enable LLM evaluations on tasks aligned with clinician needs and preferences.
Diagnostic Reasoning Prompts Reveal the Potential for Large Language Model Interpretability in Medicine
Aug 13, 2023Abstract:One of the major barriers to using large language models (LLMs) in medicine is the perception they use uninterpretable methods to make clinical decisions that are inherently different from the cognitive processes of clinicians. In this manuscript we develop novel diagnostic reasoning prompts to study whether LLMs can perform clinical reasoning to accurately form a diagnosis. We find that GPT4 can be prompted to mimic the common clinical reasoning processes of clinicians without sacrificing diagnostic accuracy. This is significant because an LLM that can use clinical reasoning to provide an interpretable rationale offers physicians a means to evaluate whether LLMs can be trusted for patient care. Novel prompting methods have the potential to expose the black box of LLMs, bringing them one step closer to safe and effective use in medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge