Thomas Gaskin
Modelling Global Trade with Optimal Transport
Sep 10, 2024Abstract:Global trade is shaped by a complex mix of factors beyond supply and demand, including tangible variables like transport costs and tariffs, as well as less quantifiable influences such as political and economic relations. Traditionally, economists model trade using gravity models, which rely on explicit covariates but often struggle to capture these subtler drivers of trade. In this work, we employ optimal transport and a deep neural network to learn a time-dependent cost function from data, without imposing a specific functional form. This approach consistently outperforms traditional gravity models in accuracy while providing natural uncertainty quantification. Applying our framework to global food and agricultural trade, we show that the global South suffered disproportionately from the war in Ukraine's impact on wheat markets. We also analyze the effects of free-trade agreements and trade disputes with China, as well as Brexit's impact on British trade with Europe, uncovering hidden patterns that trade volumes alone cannot reveal.
Neural parameter calibration and uncertainty quantification for epidemic forecasting
Dec 05, 2023


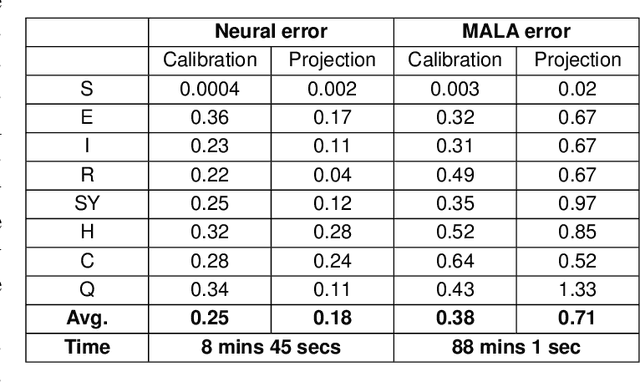
Abstract:The recent COVID-19 pandemic has thrown the importance of accurately forecasting contagion dynamics and learning infection parameters into sharp focus. At the same time, effective policy-making requires knowledge of the uncertainty on such predictions, in order, for instance, to be able to ready hospitals and intensive care units for a worst-case scenario without needlessly wasting resources. In this work, we apply a novel and powerful computational method to the problem of learning probability densities on contagion parameters and providing uncertainty quantification for pandemic projections. Using a neural network, we calibrate an ODE model to data of the spread of COVID-19 in Berlin in 2020, achieving both a significantly more accurate calibration and prediction than Markov-Chain Monte Carlo (MCMC)-based sampling schemes. The uncertainties on our predictions provide meaningful confidence intervals e.g. on infection figures and hospitalisation rates, while training and running the neural scheme takes minutes where MCMC takes hours. We show convergence of our method to the true posterior on a simplified SIR model of epidemics, and also demonstrate our method's learning capabilities on a reduced dataset, where a complex model is learned from a small number of compartments for which data is available.
Inferring networks from time series: a neural approach
Mar 30, 2023Abstract:Network structures underlie the dynamics of many complex phenomena, from gene regulation and foodwebs to power grids and social media. Yet, as they often cannot be observed directly, their connectivities must be inferred from observations of their emergent dynamics. In this work we present a powerful and fast computational method to infer large network adjacency matrices from time series data using a neural network. Using a neural network provides uncertainty quantification on the prediction in a manner that reflects both the non-convexity of the inference problem as well as the noise on the data. This is useful since network inference problems are typically underdetermined, and a feature that has hitherto been lacking from network inference methods. We demonstrate our method's capabilities by inferring line failure locations in the British power grid from observations of its response to a power cut. Since the problem is underdetermined, many classical statistical tools (e.g. regression) will not be straightforwardly applicable. Our method, in contrast, provides probability densities on each edge, allowing the use of hypothesis testing to make meaningful probabilistic statements about the location of the power cut. We also demonstrate our method's ability to learn an entire cost matrix for a non-linear model from a dataset of economic activity in Greater London. Our method outperforms OLS regression on noisy data in terms of both speed and prediction accuracy, and scales as $N^2$ where OLS is cubic. Since our technique is not specifically engineered for network inference, it represents a general parameter estimation scheme that is applicable to any parameter dimension.
Neural parameter calibration for large-scale multi-agent models
Sep 27, 2022



Abstract:Computational models have become a powerful tool in the quantitative sciences to understand the behaviour of complex systems that evolve in time. However, they often contain a potentially large number of free parameters whose values cannot be obtained from theory but need to be inferred from data. This is especially the case for models in the social sciences, economics, or computational epidemiology. Yet many current parameter estimation methods are mathematically involved and computationally slow to run. In this paper we present a computationally simple and fast method to retrieve accurate probability densities for model parameters using neural differential equations. We present a pipeline comprising multi-agent models acting as forward solvers for systems of ordinary or stochastic differential equations, and a neural network to then extract parameters from the data generated by the model. The two combined create a powerful tool that can quickly estimate densities on model parameters, even for very large systems. We demonstrate the method on synthetic time series data of the SIR model of the spread of infection, and perform an in-depth analysis of the Harris-Wilson model of economic activity on a network, representing a non-convex problem. For the latter, we apply our method both to synthetic data and to data of economic activity across Greater London. We find that our method calibrates the model orders of magnitude more accurately than a previous study of the same dataset using classical techniques, while running between 195 and 390 times faster.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge