Subhashis Hazarika
SWR-Viz: AI-assisted Interactive Visual Analytics Framework for Ship Weather Routing
Nov 19, 2025Abstract:Efficient and sustainable maritime transport increasingly depends on reliable forecasting and adaptive routing, yet operational adoption remains difficult due to forecast latencies and the need for human judgment in rapid decision-making under changing ocean conditions. We introduce SWR-Viz, an AI-assisted visual analytics framework that combines a physics-informed Fourier Neural Operator wave forecast model with SIMROUTE-based routing and interactive emissions analytics. The framework generates near-term forecasts directly from current conditions, supports data assimilation with sparse observations, and enables rapid exploration of what-if routing scenarios. We evaluate the forecast models and SWR-Viz framework along key shipping corridors in the Japan Coast and Gulf of Mexico, showing both improved forecast stability and realistic routing outcomes comparable to ground-truth reanalysis wave products. Expert feedback highlights the usability of SWR-Viz, its ability to isolate voyage segments with high emission reduction potential, and its value as a practical decision-support system. More broadly, this work illustrates how lightweight AI forecasting can be integrated with interactive visual analytics to support human-centered decision-making in complex geospatial and environmental domains.
Simultaneous emulation and downscaling with physically-consistent deep learning-based regional ocean emulators
Jan 09, 2025



Abstract:Building on top of the success in AI-based atmospheric emulation, we propose an AI-based ocean emulation and downscaling framework focusing on the high-resolution regional ocean over Gulf of Mexico. Regional ocean emulation presents unique challenges owing to the complex bathymetry and lateral boundary conditions as well as from fundamental biases in deep learning-based frameworks, such as instability and hallucinations. In this paper, we develop a deep learning-based framework to autoregressively integrate ocean-surface variables over the Gulf of Mexico at $8$ Km spatial resolution without unphysical drifts over decadal time scales and simulataneously downscale and bias-correct it to $4$ Km resolution using a physics-constrained generative model. The framework shows both short-term skills as well as accurate long-term statistics in terms of mean and variability.
Are Generative AI systems Capable of Supporting Information Needs of Patients?
Jan 31, 2024Abstract:Patients managing a complex illness such as cancer face a complex information challenge where they not only must learn about their illness but also how to manage it. Close interaction with healthcare experts (radiologists, oncologists) can improve patient learning and thereby, their disease outcome. However, this approach is resource intensive and takes expert time away from other critical tasks. Given the recent advancements in Generative AI models aimed at improving the healthcare system, our work investigates whether and how generative visual question answering systems can responsibly support patient information needs in the context of radiology imaging data. We conducted a formative need-finding study in which participants discussed chest computed tomography (CT) scans and associated radiology reports of a fictitious close relative with a cardiothoracic radiologist. Using thematic analysis of the conversation between participants and medical experts, we identified commonly occurring themes across interactions, including clarifying medical terminology, locating the problems mentioned in the report in the scanned image, understanding disease prognosis, discussing the next diagnostic steps, and comparing treatment options. Based on these themes, we evaluated two state-of-the-art generative visual language models against the radiologist's responses. Our results reveal variability in the quality of responses generated by the models across various themes. We highlight the importance of patient-facing generative AI systems to accommodate a diverse range of conversational themes, catering to the real-world informational needs of patients.
HAiVA: Hybrid AI-assisted Visual Analysis Framework to Study the Effects of Cloud Properties on Climate Patterns
May 13, 2023

Abstract:Clouds have a significant impact on the Earth's climate system. They play a vital role in modulating Earth's radiation budget and driving regional changes in temperature and precipitation. This makes clouds ideal for climate intervention techniques like Marine Cloud Brightening (MCB) which refers to modification in cloud reflectivity, thereby cooling the surrounding region. However, to avoid unintended effects of MCB, we need a better understanding of the complex cloud to climate response function. Designing and testing such interventions scenarios with conventional Earth System Models is computationally expensive. Therefore, we propose a hybrid AI-assisted visual analysis framework to drive such scientific studies and facilitate interactive what-if investigation of different MCB intervention scenarios to assess their intended and unintended impacts on climate patterns. We work with a team of climate scientists to develop a suite of hybrid AI models emulating cloud-climate response function and design a tightly coupled frontend interactive visual analysis system to perform different MCB intervention experiments.
Climate Intervention Analysis using AI Model Guided by Statistical Physics Principles
Feb 07, 2023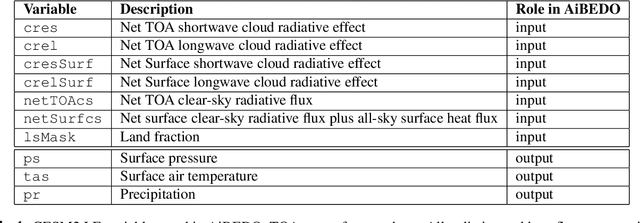
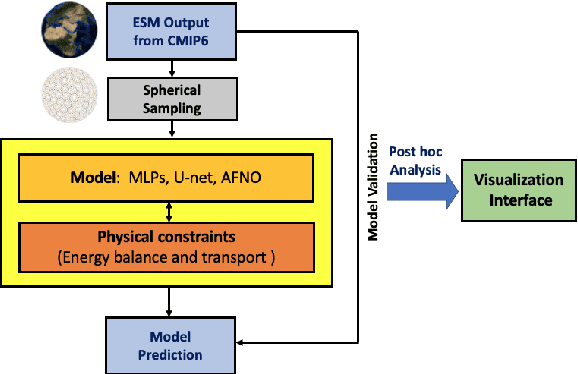
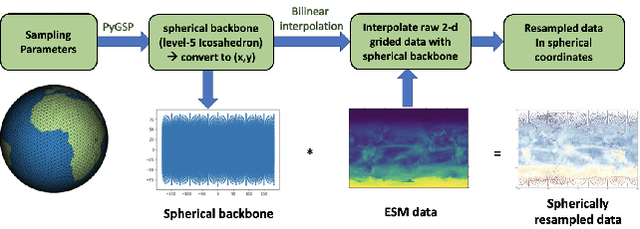
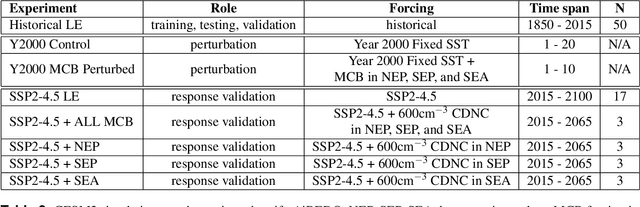
Abstract:The availability of training data remains a significant obstacle for the implementation of machine learning in scientific applications. In particular, estimating how a system might respond to external forcings or perturbations requires specialized labeled data or targeted simulations, which may be computationally intensive to generate at scale. In this study, we propose a novel solution to this challenge by utilizing a principle from statistical physics known as the Fluctuation-Dissipation Theorem (FDT) to discover knowledge using an AI model that can rapidly produce scenarios for different external forcings. By leveraging FDT, we are able to extract information encoded in a large dataset produced by Earth System Models, which includes 8250 years of internal climate fluctuations, to estimate the climate system's response to forcings. Our model, AiBEDO, is capable of capturing the complex, multi-timescale effects of radiation perturbations on global and regional surface climate, allowing for a substantial acceleration of the exploration of the impacts of spatially-heterogenous climate forcers. To demonstrate the utility of AiBEDO, we use the example of a climate intervention technique called Marine Cloud Brightening, with the ultimate goal of optimizing the spatial pattern of cloud brightening to achieve regional climate targets and prevent known climate tipping points. While we showcase the effectiveness of our approach in the context of climate science, it is generally applicable to other scientific disciplines that are limited by the extensive computational demands of domain simulation models. Source code of AiBEDO framework is made available at https://github.com/kramea/kdd_aibedo. A sample dataset is made available at https://doi.org/10.5281/zenodo.7597027. Additional data available upon request.
Accelerating exploration of Marine Cloud Brightening impacts on tipping points Using an AI Implementation of Fluctuation-Dissipation Theorem
Feb 03, 2023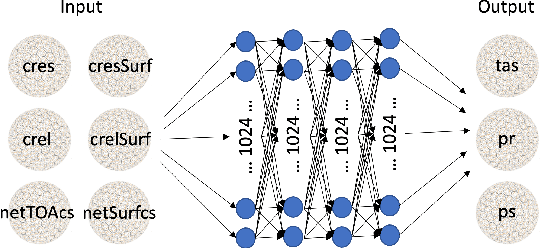
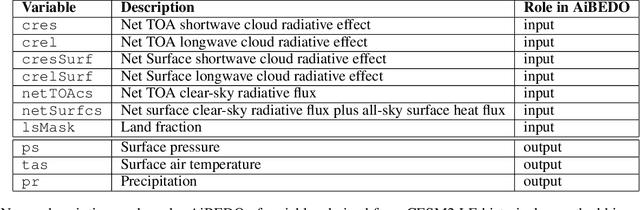
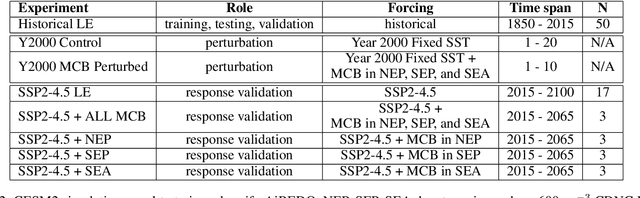
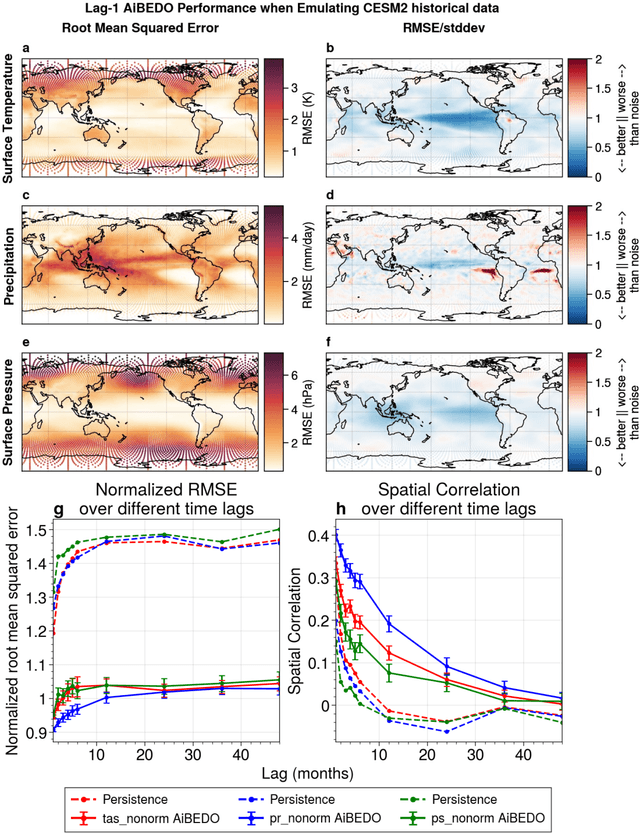
Abstract:Marine cloud brightening (MCB) is a proposed climate intervention technology to partially offset greenhouse gas warming and possibly avoid crossing climate tipping points. The impacts of MCB on regional climate are typically estimated using computationally expensive Earth System Model (ESM) simulations, preventing a thorough assessment of the large possibility space of potential MCB interventions. Here, we describe an AI model, named AiBEDO, that can be used to rapidly projects climate responses to forcings via a novel application of the Fluctuation-Dissipation Theorem (FDT). AiBEDO is a Multilayer Perceptron (MLP) model that uses maps monthly-mean radiation anomalies to surface climate anomalies at a range of time lags. By leveraging a large existing dataset of ESM simulations containing internal climate noise, we use AiBEDO to construct an FDT operator that successfully projects climate responses to MCB forcing, when evaluated against ESM simulations. We propose that AiBEDO-FDT can be used to optimize MCB forcing patterns to reduce tipping point risks while minimizing negative side effects in other parts of the climate.
Relationship-aware Multivariate Sampling Strategy for Scientific Simulation Data
Aug 31, 2020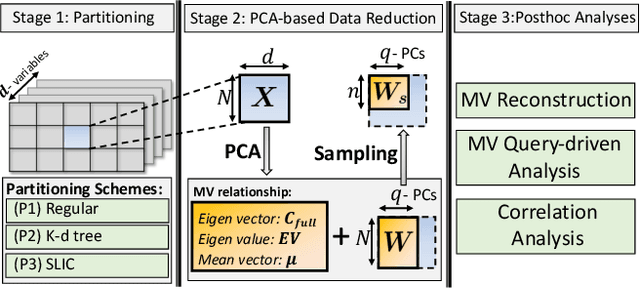
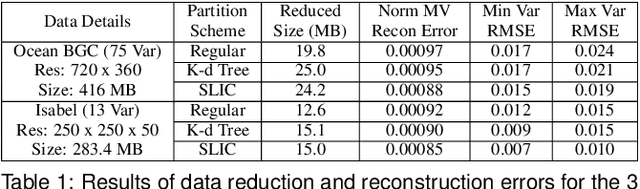
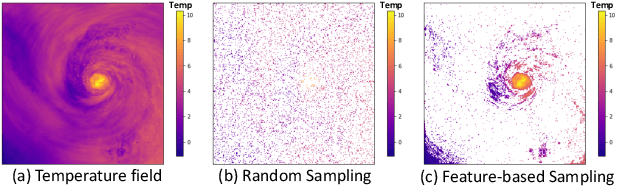

Abstract:With the increasing computational power of current supercomputers, the size of data produced by scientific simulations is rapidly growing. To reduce the storage footprint and facilitate scalable post-hoc analyses of such scientific data sets, various data reduction/summarization methods have been proposed over the years. Different flavors of sampling algorithms exist to sample the high-resolution scientific data, while preserving important data properties required for subsequent analyses. However, most of these sampling algorithms are designed for univariate data and cater to post-hoc analyses of single variables. In this work, we propose a multivariate sampling strategy which preserves the original variable relationships and enables different multivariate analyses directly on the sampled data. Our proposed strategy utilizes principal component analysis to capture the variance of multivariate data and can be built on top of any existing state-of-the-art sampling algorithms for single variables. In addition, we also propose variants of different data partitioning schemes (regular and irregular) to efficiently model the local multivariate relationships. Using two real-world multivariate data sets, we demonstrate the efficacy of our proposed multivariate sampling strategy with respect to its data reduction capabilities as well as the ease of performing efficient post-hoc multivariate analyses.
Deep Learning-Based Feature-Aware Data Modeling for Complex Physics Simulations
Dec 08, 2019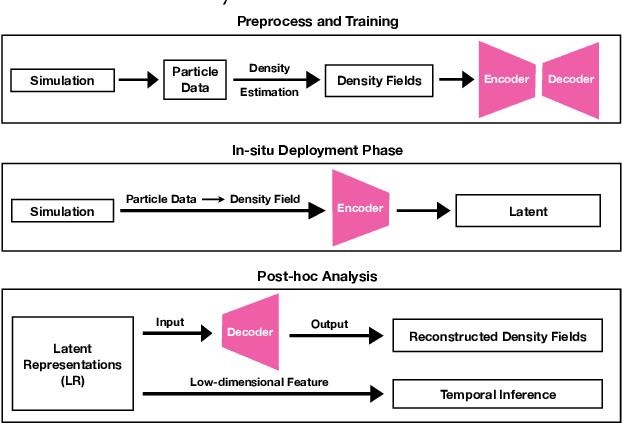
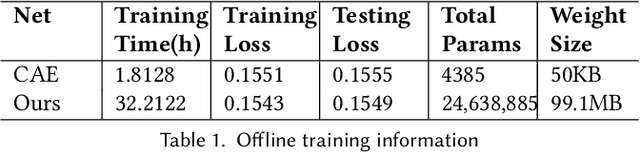
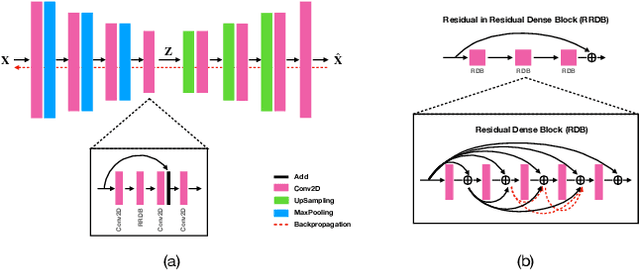

Abstract:Data modeling and reduction for in situ is important. Feature-driven methods for in situ data analysis and reduction are a priority for future exascale machines as there are currently very few such methods. We investigate a deep-learning based workflow that targets in situ data processing using autoencoders. We propose a Residual Autoencoder integrated Residual in Residual Dense Block (RRDB) to obtain better performance. Our proposed framework compressed our test data into 66 KB from 2.1 MB per 3D volume timestep.
NNVA: Neural Network Assisted Visual Analysis of Yeast Cell Polarization Simulation
Apr 19, 2019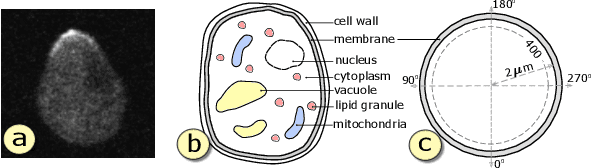
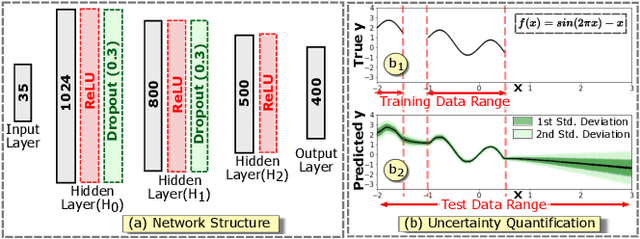
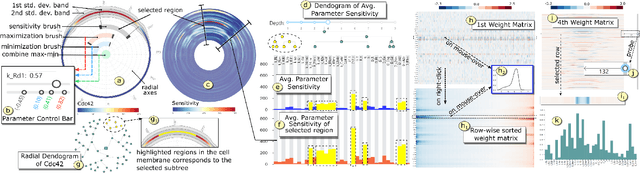
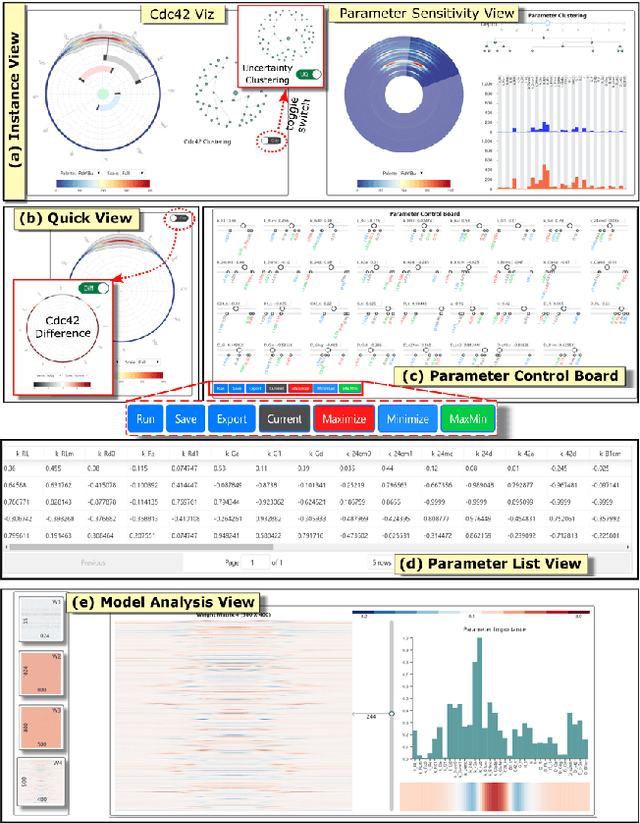
Abstract:Complex computational models are often designed to simulate real-world physical phenomena in many scientific disciplines. However, these simulation models tend to be computationally very expensive and involve a large number of simulation input parameters which need to be analyzed and properly calibrated before the models can be applied for real scientific studies. We propose a visual analysis system to facilitate interactive exploratory analysis of high-dimensional input parameter space for a complex yeast cell polarization simulation. The proposed system can assist the computational biologists, who designed the simulation model, to visually calibrate the input parameters by modifying the parameter values and immediately visualizing the predicted simulation outcome without having the need to run the original expensive simulation for every instance. Our proposed visual analysis system is driven by a trained neural network-based surrogate model as the backend analysis framework. Surrogate models are widely used in the field of simulation sciences to efficiently analyze computationally expensive simulation models. In this work, we demonstrate the advantage of using neural networks as surrogate models for visual analysis by incorporating some of the recent advances in the field of uncertainty quantification, interpretability and explainability of neural network-based models. We utilize the trained network to perform interactive parameter sensitivity analysis of the original simulation at multiple levels-of-detail as well as recommend optimal parameter configurations using the activation maximization framework of neural networks. We also facilitate detail analysis of the trained network to extract useful insights about the simulation model, learned by the network, during the training process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge