Stephen M. Gordon
Decoding Neural Activity to Assess Individual Latent State in Ecologically Valid Contexts
Apr 18, 2023Abstract:There exist very few ways to isolate cognitive processes, historically defined via highly controlled laboratory studies, in more ecologically valid contexts. Specifically, it remains unclear as to what extent patterns of neural activity observed under such constraints actually manifest outside the laboratory in a manner that can be used to make an accurate inference about the latent state, associated cognitive process, or proximal behavior of the individual. Improving our understanding of when and how specific patterns of neural activity manifest in ecologically valid scenarios would provide validation for laboratory-based approaches that study similar neural phenomena in isolation and meaningful insight into the latent states that occur during complex tasks. We argue that domain generalization methods from the brain-computer interface community have the potential to address this challenge. We previously used such an approach to decode phasic neural responses associated with visual target discrimination. Here, we extend that work to more tonic phenomena such as internal latent states. We use data from two highly controlled laboratory paradigms to train two separate domain-generalized models. We apply the trained models to an ecologically valid paradigm in which participants performed multiple, concurrent driving-related tasks. Using the pretrained models, we derive estimates of the underlying latent state and associated patterns of neural activity. Importantly, as the patterns of neural activity change along the axis defined by the original training data, we find changes in behavior and task performance consistent with the observations from the original, laboratory paradigms. We argue that these results lend ecological validity to those experimental designs and provide a methodology for understanding the relationship between observed neural activity and behavior during complex tasks.
2021 BEETL Competition: Advancing Transfer Learning for Subject Independence & Heterogenous EEG Data Sets
Feb 14, 2022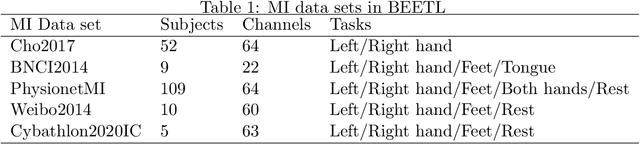
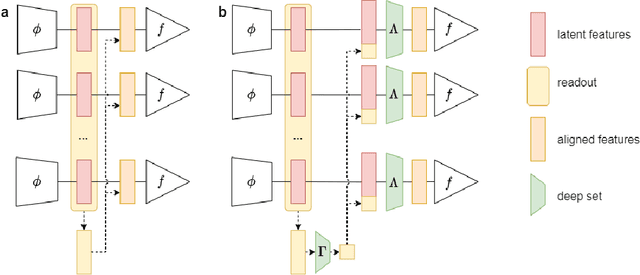
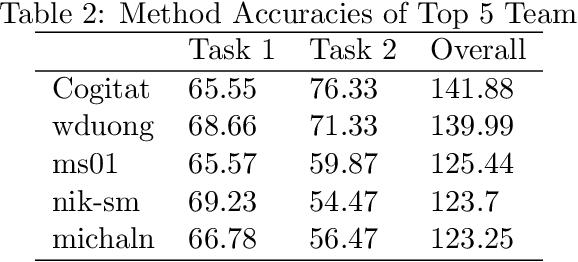
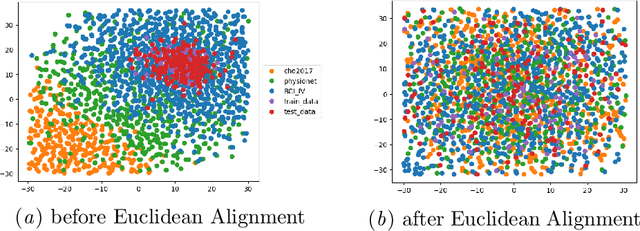
Abstract:Transfer learning and meta-learning offer some of the most promising avenues to unlock the scalability of healthcare and consumer technologies driven by biosignal data. This is because current methods cannot generalise well across human subjects' data and handle learning from different heterogeneously collected data sets, thus limiting the scale of training data. On the other side, developments in transfer learning would benefit significantly from a real-world benchmark with immediate practical application. Therefore, we pick electroencephalography (EEG) as an exemplar for what makes biosignal machine learning hard. We design two transfer learning challenges around diagnostics and Brain-Computer-Interfacing (BCI), that have to be solved in the face of low signal-to-noise ratios, major variability among subjects, differences in the data recording sessions and techniques, and even between the specific BCI tasks recorded in the dataset. Task 1 is centred on the field of medical diagnostics, addressing automatic sleep stage annotation across subjects. Task 2 is centred on Brain-Computer Interfacing (BCI), addressing motor imagery decoding across both subjects and data sets. The BEETL competition with its over 30 competing teams and its 3 winning entries brought attention to the potential of deep transfer learning and combinations of set theory and conventional machine learning techniques to overcome the challenges. The results set a new state-of-the-art for the real-world BEETL benchmark.
EEGNet: A Compact Convolutional Network for EEG-based Brain-Computer Interfaces
May 16, 2018
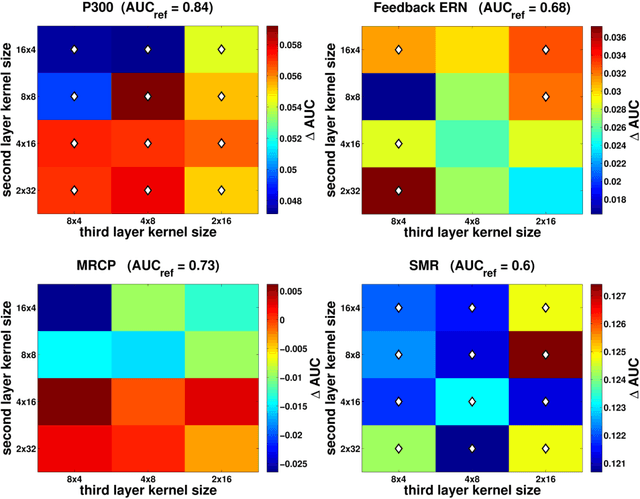
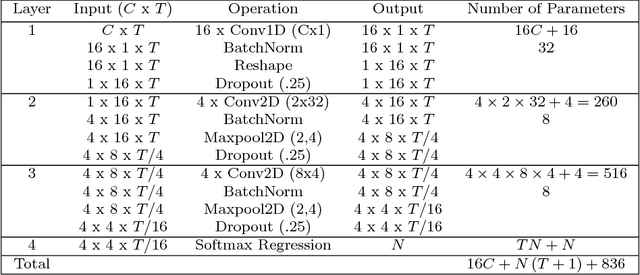
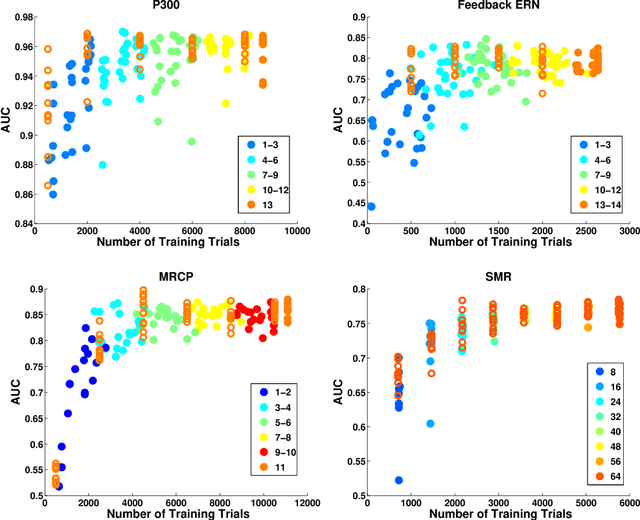
Abstract:Brain computer interfaces (BCI) enable direct communication with a computer, using neural activity as the control signal. This neural signal is generally chosen from a variety of well-studied electroencephalogram (EEG) signals. For a given BCI paradigm, feature extractors and classifiers are tailored to the distinct characteristics of its expected EEG control signal, limiting its application to that specific signal. Convolutional Neural Networks (CNNs), which have been used in computer vision and speech recognition, have successfully been applied to EEG-based BCIs; however, they have mainly been applied to single BCI paradigms and thus it remains unclear how these architectures generalize to other paradigms. Here, we ask if we can design a single CNN architecture to accurately classify EEG signals from different BCI paradigms, while simultaneously being as compact as possible. In this work we introduce EEGNet, a compact convolutional network for EEG-based BCIs. We introduce the use of depthwise and separable convolutions to construct an EEG-specific model which encapsulates well-known EEG feature extraction concepts for BCI. We compare EEGNet to current state-of-the-art approaches across four BCI paradigms: P300 visual-evoked potentials, error-related negativity responses (ERN), movement-related cortical potentials (MRCP), and sensory motor rhythms (SMR). We show that EEGNet generalizes across paradigms better than the reference algorithms when only limited training data is available. We demonstrate three different approaches to visualize the contents of a trained EEGNet model to enable interpretation of the learned features. Our results suggest that EEGNet is robust enough to learn a wide variety of interpretable features over a range of BCI tasks, suggesting that the observed performances were not due to artifact or noise sources in the data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge