Stephen H. Muggleton
Boolean Matrix Logic Programming
Aug 19, 2024Abstract:We describe a datalog query evaluation approach based on efficient and composable boolean matrix manipulation modules. We first define an overarching problem, Boolean Matrix Logic Programming (BMLP), which uses boolean matrices as an alternative computation to evaluate datalog programs. We develop two novel BMLP modules for bottom-up inferences on linear dyadic recursive datalog programs, and show how additional modules can extend this capability to compute both linear and non-linear recursive datalog programs of arity two. Our empirical results demonstrate that these modules outperform general-purpose and specialised systems by factors of 30x and 9x, respectively, when evaluating large programs with millions of facts. This boolean matrix approach significantly enhances the efficiency of datalog querying to support logic programming techniques.
Active learning of digenic functions with boolean matrix logic programming
Aug 19, 2024Abstract:We apply logic-based machine learning techniques to facilitate cellular engineering and drive biological discovery, based on comprehensive databases of metabolic processes called genome-scale metabolic network models (GEMs). Predicted host behaviours are not always correctly described by GEMs. Learning the intricate genetic interactions within GEMs presents computational and empirical challenges. To address these, we describe a novel approach called Boolean Matrix Logic Programming (BMLP) by leveraging boolean matrices to evaluate large logic programs. We introduce a new system, $BMLP_{active}$, which efficiently explores the genomic hypothesis space by guiding informative experimentation through active learning. In contrast to sub-symbolic methods, $BMLP_{active}$ encodes a state-of-the-art GEM of a widely accepted bacterial host in an interpretable and logical representation using datalog logic programs. Notably, $BMLP_{active}$ can successfully learn the interaction between a gene pair with fewer training examples than random experimentation, overcoming the increase in experimental design space. $BMLP_{active}$ enables rapid optimisation of metabolic models and offers a realistic approach to a self-driving lab for microbial engineering.
Simulating Petri nets with Boolean Matrix Logic Programming
May 18, 2024Abstract:Recent attention to relational knowledge bases has sparked a demand for understanding how relations change between entities. Petri nets can represent knowledge structure and dynamically simulate interactions between entities, and thus they are well suited for achieving this goal. However, logic programs struggle to deal with extensive Petri nets due to the limitations of high-level symbol manipulations. To address this challenge, we introduce a novel approach called Boolean Matrix Logic Programming (BMLP), utilising boolean matrices as an alternative computation mechanism for Prolog to evaluate logic programs. Within this framework, we propose two novel BMLP algorithms for simulating a class of Petri nets known as elementary nets. This is done by transforming elementary nets into logically equivalent datalog programs. We demonstrate empirically that BMLP algorithms can evaluate these programs 40 times faster than tabled B-Prolog, SWI-Prolog, XSB-Prolog and Clingo. Our work enables the efficient simulation of elementary nets using Prolog, expanding the scope of analysis, learning and verification of complex systems with logic programming techniques.
Boolean matrix logic programming for active learning of gene functions in genome-scale metabolic network models
May 10, 2024Abstract:Techniques to autonomously drive research have been prominent in Computational Scientific Discovery, while Synthetic Biology is a field of science that focuses on designing and constructing new biological systems for useful purposes. Here we seek to apply logic-based machine learning techniques to facilitate cellular engineering and drive biological discovery. Comprehensive databases of metabolic processes called genome-scale metabolic network models (GEMs) are often used to evaluate cellular engineering strategies to optimise target compound production. However, predicted host behaviours are not always correctly described by GEMs, often due to errors in the models. The task of learning the intricate genetic interactions within GEMs presents computational and empirical challenges. To address these, we describe a novel approach called Boolean Matrix Logic Programming (BMLP) by leveraging boolean matrices to evaluate large logic programs. We introduce a new system, $BMLP_{active}$, which efficiently explores the genomic hypothesis space by guiding informative experimentation through active learning. In contrast to sub-symbolic methods, $BMLP_{active}$ encodes a state-of-the-art GEM of a widely accepted bacterial host in an interpretable and logical representation using datalog logic programs. Notably, $BMLP_{active}$ can successfully learn the interaction between a gene pair with fewer training examples than random experimentation, overcoming the increase in experimental design space. $BMLP_{active}$ enables rapid optimisation of metabolic models to reliably engineer biological systems for producing useful compounds. It offers a realistic approach to creating a self-driving lab for microbial engineering.
Human Comprehensible Active Learning of Genome-Scale Metabolic Networks
Aug 31, 2023
Abstract:An important application of Synthetic Biology is the engineering of the host cell system to yield useful products. However, an increase in the scale of the host system leads to huge design space and requires a large number of validation trials with high experimental costs. A comprehensible machine learning approach that efficiently explores the hypothesis space and guides experimental design is urgently needed for the Design-Build-Test-Learn (DBTL) cycle of the host cell system. We introduce a novel machine learning framework ILP-iML1515 based on Inductive Logic Programming (ILP) that performs abductive logical reasoning and actively learns from training examples. In contrast to numerical models, ILP-iML1515 is built on comprehensible logical representations of a genome-scale metabolic model and can update the model by learning new logical structures from auxotrophic mutant trials. The ILP-iML1515 framework 1) allows high-throughput simulations and 2) actively selects experiments that reduce the experimental cost of learning gene functions in comparison to randomly selected experiments.
Explanatory machine learning for sequential human teaching
May 20, 2022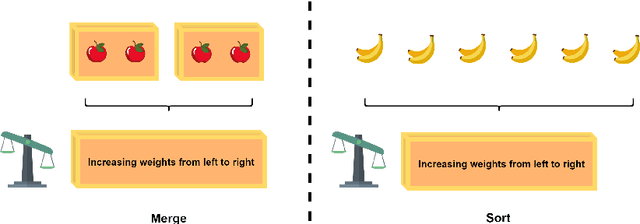

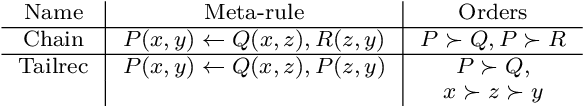
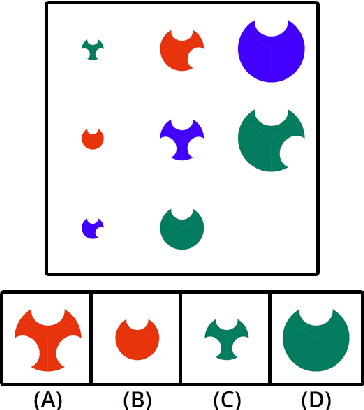
Abstract:The topic of comprehensibility of machine-learned theories has recently drawn increasing attention. Inductive Logic Programming (ILP) uses logic programming to derive logic theories from small data based on abduction and induction techniques. Learned theories are represented in the form of rules as declarative descriptions of obtained knowledge. In earlier work, the authors provided the first evidence of a measurable increase in human comprehension based on machine-learned logic rules for simple classification tasks. In a later study, it was found that the presentation of machine-learned explanations to humans can produce both beneficial and harmful effects in the context of game learning. We continue our investigation of comprehensibility by examining the effects of the ordering of concept presentations on human comprehension. In this work, we examine the explanatory effects of curriculum order and the presence of machine-learned explanations for sequential problem-solving. We show that 1) there exist tasks A and B such that learning A before B has a better human comprehension with respect to learning B before A and 2) there exist tasks A and B such that the presence of explanations when learning A contributes to improved human comprehension when subsequently learning B. We propose a framework for the effects of sequential teaching on comprehension based on an existing definition of comprehensibility and provide evidence for support from data collected in human trials. Empirical results show that sequential teaching of concepts with increasing complexity a) has a beneficial effect on human comprehension and b) leads to human re-discovery of divide-and-conquer problem-solving strategies, and c) studying machine-learned explanations allows adaptations of human problem-solving strategy with better performance.
Meta-Interpretive Learning as Metarule Specialisation
Jun 09, 2021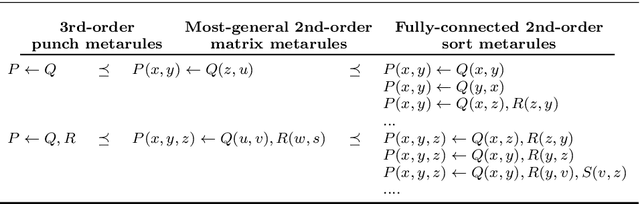
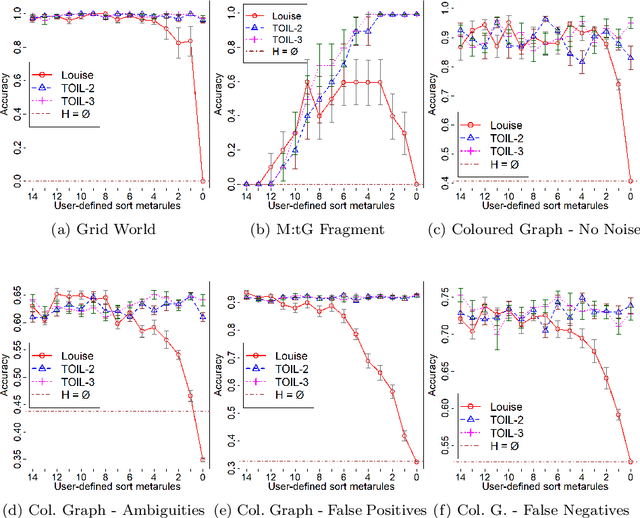
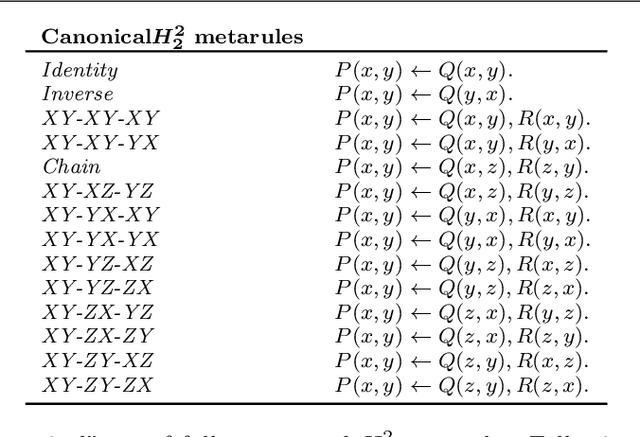
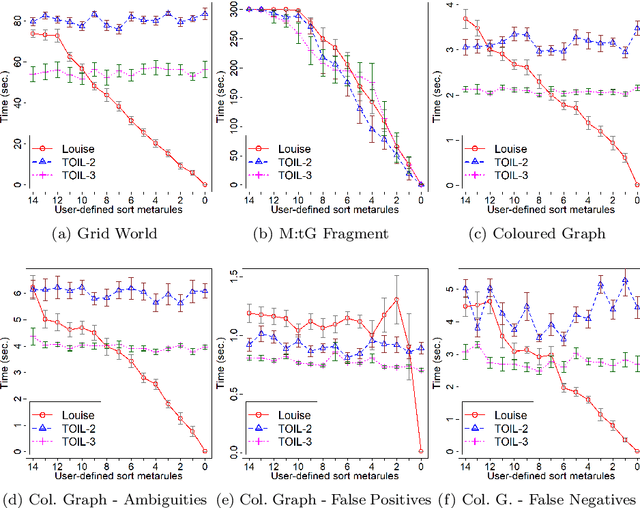
Abstract:In Meta-Interpretive Learning (MIL) the metarules, second-order datalog clauses acting as inductive bias, are manually defined by the user. In this work we show that second-order metarules for MIL can be learned by MIL. We define a generality ordering of metarules by $\theta$-subsumption and show that user-defined sort metarules are derivable by specialisation of the most-general matrix metarules in a language class; and that these matrix metarules are in turn derivable by specialisation of third-order punch metarules with variables that range over the set of second-order literals and for which only an upper bound on their number of literals need be user-defined. We show that the cardinality of a metarule language is polynomial in the number of literals in punch metarules. We re-frame MIL as metarule specialisation by resolution. We modify the MIL metarule specialisation operator to return new metarules rather than first-order clauses and prove the correctness of the new operator. We implement the new operator as TOIL, a sub-system of the MIL system Louise. Our experiments show that as user-defined sort metarules are progressively replaced by sort metarules learned by TOIL, Louise's predictive accuracy is maintained at the cost of a small increase in training times. We conclude that automatically derived metarules can replace user-defined metarules.
Automated Biodesign Engineering by Abductive Meta-Interpretive Learning
May 17, 2021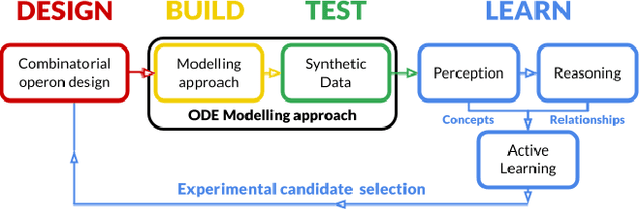
Abstract:The application of Artificial Intelligence (AI) to synthetic biology will provide the foundation for the creation of a high throughput automated platform for genetic design, in which a learning machine is used to iteratively optimise the system through a design-build-test-learn (DBTL) cycle. However, mainstream machine learning techniques represented by deep learning lacks the capability to represent relational knowledge and requires prodigious amounts of annotated training data. These drawbacks strongly restrict AI's role in synthetic biology in which experimentation is inherently resource and time intensive. In this work, we propose an automated biodesign engineering framework empowered by Abductive Meta-Interpretive Learning ($Meta_{Abd}$), a novel machine learning approach that combines symbolic and sub-symbolic machine learning, to further enhance the DBTL cycle by enabling the learning machine to 1) exploit domain knowledge and learn human-interpretable models that are expressed by formal languages such as first-order logic; 2) simultaneously optimise the structure and parameters of the models to make accurate numerical predictions; 3) reduce the cost of experiments and effort on data annotation by actively generating hypotheses and examples. To verify the effectiveness of $Meta_{Abd}$, we have modelled a synthetic dataset for the production of proteins from a three gene operon in a microbial host, which represents a common synthetic biology problem.
Inductive logic programming at 30
Feb 21, 2021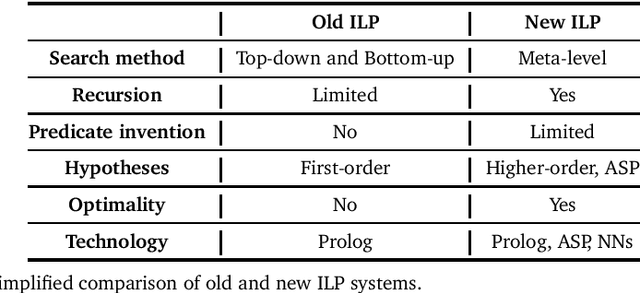
Abstract:Inductive logic programming (ILP) is a form of logic-based machine learning. The goal of ILP is to induce a hypothesis (a logic program) that generalises given training examples and background knowledge. As ILP turns 30, we survey recent work in the field. In this survey, we focus on (i) new meta-level search methods, (ii) techniques for learning recursive programs that generalise from few examples, (iii) new approaches for predicate invention, and (iv) the use of different technologies, notably answer set programming and neural networks. We conclude by discussing some of the current limitations of ILP and discuss directions for future research.
Top Program Construction and Reduction for polynomial time Meta-Interpretive Learning
Jan 13, 2021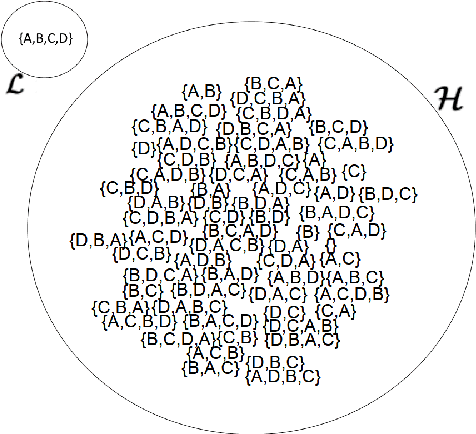
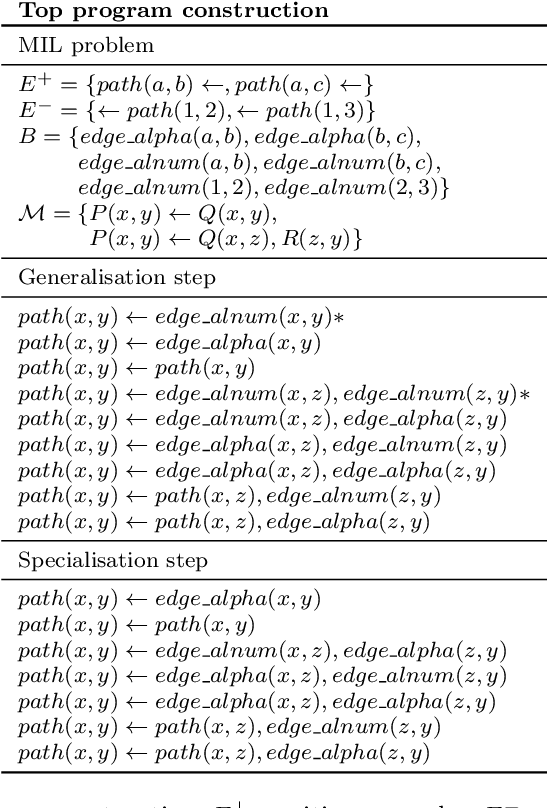
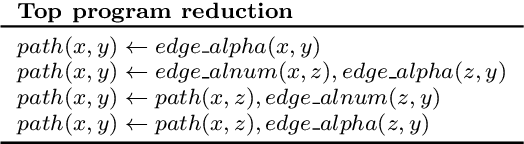
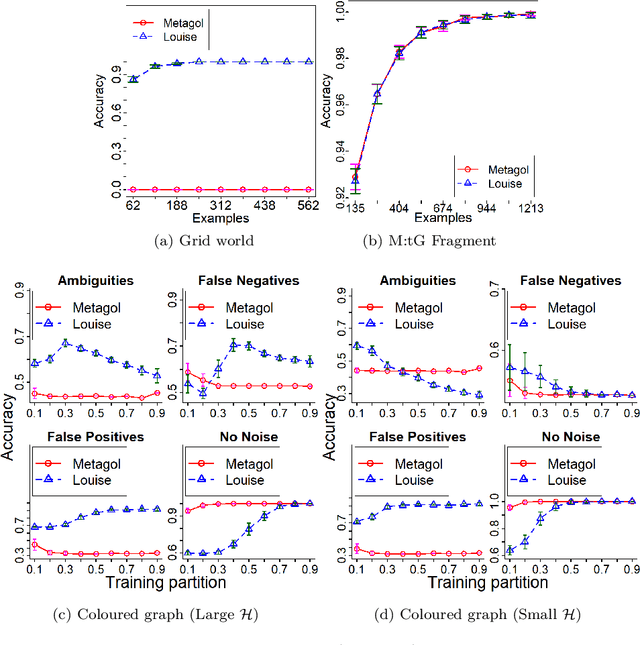
Abstract:Meta-Interpretive Learners, like most ILP systems, learn by searching for a correct hypothesis in the hypothesis space, the powerset of all constructible clauses. We show how this exponentially-growing search can be replaced by the construction of a Top program: the set of clauses in all correct hypotheses that is itself a correct hypothesis. We give an algorithm for Top program construction and show that it constructs a correct Top program in polynomial time and from a finite number of examples. We implement our algorithm in Prolog as the basis of a new MIL system, Louise, that constructs a Top program and then reduces it by removing redundant clauses. We compare Louise to the state-of-the-art search-based MIL system Metagol in experiments on grid world navigation, graph connectedness and grammar learning datasets and find that Louise improves on Metagol's predictive accuracy when the hypothesis space and the target theory are both large, or when the hypothesis space does not include a correct hypothesis because of "classification noise" in the form of mislabelled examples. When the hypothesis space or the target theory are small, Louise and Metagol perform equally well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge