Liam Hallett
Human Comprehensible Active Learning of Genome-Scale Metabolic Networks
Aug 31, 2023
Abstract:An important application of Synthetic Biology is the engineering of the host cell system to yield useful products. However, an increase in the scale of the host system leads to huge design space and requires a large number of validation trials with high experimental costs. A comprehensible machine learning approach that efficiently explores the hypothesis space and guides experimental design is urgently needed for the Design-Build-Test-Learn (DBTL) cycle of the host cell system. We introduce a novel machine learning framework ILP-iML1515 based on Inductive Logic Programming (ILP) that performs abductive logical reasoning and actively learns from training examples. In contrast to numerical models, ILP-iML1515 is built on comprehensible logical representations of a genome-scale metabolic model and can update the model by learning new logical structures from auxotrophic mutant trials. The ILP-iML1515 framework 1) allows high-throughput simulations and 2) actively selects experiments that reduce the experimental cost of learning gene functions in comparison to randomly selected experiments.
Automated Biodesign Engineering by Abductive Meta-Interpretive Learning
May 17, 2021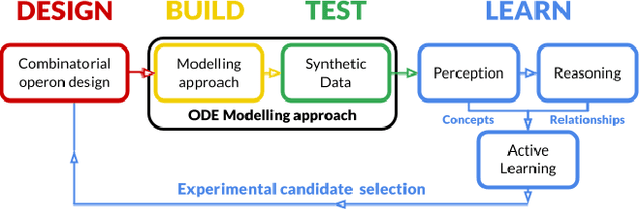
Abstract:The application of Artificial Intelligence (AI) to synthetic biology will provide the foundation for the creation of a high throughput automated platform for genetic design, in which a learning machine is used to iteratively optimise the system through a design-build-test-learn (DBTL) cycle. However, mainstream machine learning techniques represented by deep learning lacks the capability to represent relational knowledge and requires prodigious amounts of annotated training data. These drawbacks strongly restrict AI's role in synthetic biology in which experimentation is inherently resource and time intensive. In this work, we propose an automated biodesign engineering framework empowered by Abductive Meta-Interpretive Learning ($Meta_{Abd}$), a novel machine learning approach that combines symbolic and sub-symbolic machine learning, to further enhance the DBTL cycle by enabling the learning machine to 1) exploit domain knowledge and learn human-interpretable models that are expressed by formal languages such as first-order logic; 2) simultaneously optimise the structure and parameters of the models to make accurate numerical predictions; 3) reduce the cost of experiments and effort on data annotation by actively generating hypotheses and examples. To verify the effectiveness of $Meta_{Abd}$, we have modelled a synthetic dataset for the production of proteins from a three gene operon in a microbial host, which represents a common synthetic biology problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge