Solha Kang
Detecting Regional Spurious Correlations in Vision Transformers via Token Discarding
Sep 04, 2025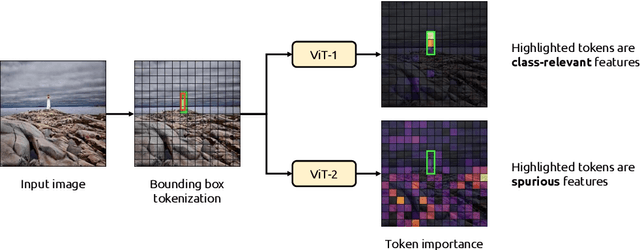


Abstract:Due to their powerful feature association capabilities, neural network-based computer vision models have the ability to detect and exploit unintended patterns within the data, potentially leading to correct predictions based on incorrect or unintended but statistically relevant signals. These clues may vary from simple color aberrations to small texts within the image. In situations where these unintended signals align with the predictive task, models can mistakenly link these features with the task and rely on them for making predictions. This phenomenon is referred to as spurious correlations, where patterns appear to be associated with the task but are actually coincidental. As a result, detection and mitigation of spurious correlations have become crucial tasks for building trustworthy, reliable, and generalizable machine learning models. In this work, we present a novel method to detect spurious correlations in vision transformers, a type of neural network architecture that gained significant popularity in recent years. Using both supervised and self-supervised trained models, we present large-scale experiments on the ImageNet dataset demonstrating the ability of the proposed method to identify spurious correlations. We also find that, even if the same architecture is used, the training methodology has a significant impact on the model's reliance on spurious correlations. Furthermore, we show that certain classes in the ImageNet dataset contain spurious signals that are easily detected by the models and discuss the underlying reasons for those spurious signals. In light of our findings, we provide an exhaustive list of the aforementioned images and call for caution in their use in future research efforts. Lastly, we present a case study investigating spurious signals in invasive breast mass classification, grounding our work in real-world scenarios.
Exploring Patient Data Requirements in Training Effective AI Models for MRI-based Breast Cancer Classification
Feb 22, 2025Abstract:The past decade has witnessed a substantial increase in the number of startups and companies offering AI-based solutions for clinical decision support in medical institutions. However, the critical nature of medical decision-making raises several concerns about relying on external software. Key issues include potential variations in image modalities and the medical devices used to obtain these images, potential legal issues, and adversarial attacks. Fortunately, the open-source nature of machine learning research has made foundation models publicly available and straightforward to use for medical applications. This accessibility allows medical institutions to train their own AI-based models, thereby mitigating the aforementioned concerns. Given this context, an important question arises: how much data do medical institutions need to train effective AI models? In this study, we explore this question in relation to breast cancer detection, a particularly contested area due to the prevalence of this disease, which affects approximately 1 in every 8 women. Through large-scale experiments on various patient sizes in the training set, we show that medical institutions do not need a decade's worth of MRI images to train an AI model that performs competitively with the state-of-the-art, provided the model leverages foundation models. Furthermore, we observe that for patient counts greater than 50, the number of patients in the training set has a negligible impact on the performance of models and that simple ensembles further improve the results without additional complexity.
Identifying Critical Tokens for Accurate Predictions in Transformer-based Medical Imaging Models
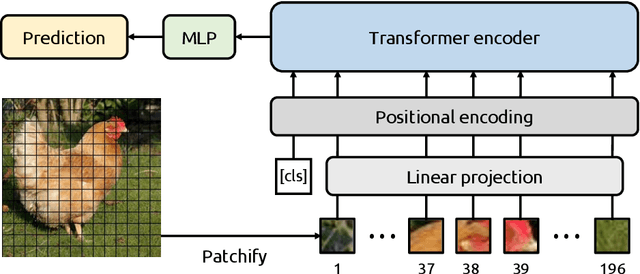
Jan 26, 2025Abstract:With the advancements in self-supervised learning (SSL), transformer-based computer vision models have recently demonstrated superior results compared to convolutional neural networks (CNNs) and are poised to dominate the field of artificial intelligence (AI)-based medical imaging in the upcoming years. Nevertheless, similar to CNNs, unveiling the decision-making process of transformer-based models remains a challenge. In this work, we take a step towards demystifying the decision-making process of transformer-based medical imaging models and propose Token Insight, a novel method that identifies the critical tokens that contribute to the prediction made by the model. Our method relies on the principled approach of token discarding native to transformer-based models, requires no additional module, and can be applied to any transformer model. Using the proposed approach, we quantify the importance of each token based on its contribution to the prediction and enable a more nuanced understanding of the model's decisions. Our experimental results which are showcased on the problem of colonic polyp identification using both supervised and self-supervised pretrained vision transformers indicate that Token Insight contributes to a more transparent and interpretable transformer-based medical imaging model, fostering trust and facilitating broader adoption in clinical settings.
Self-supervised Benchmark Lottery on ImageNet: Do Marginal Improvements Translate to Improvements on Similar Datasets?
Jan 26, 2025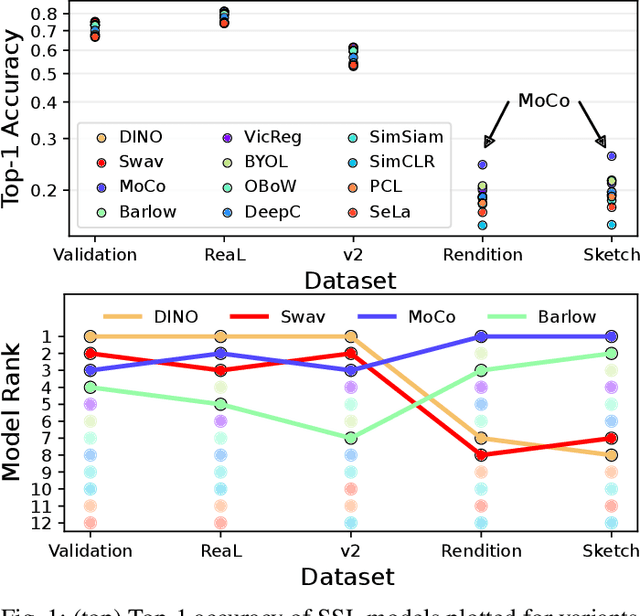
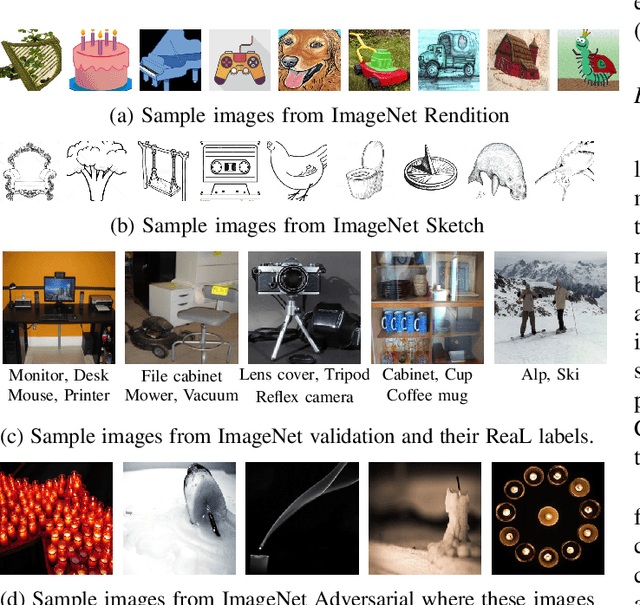
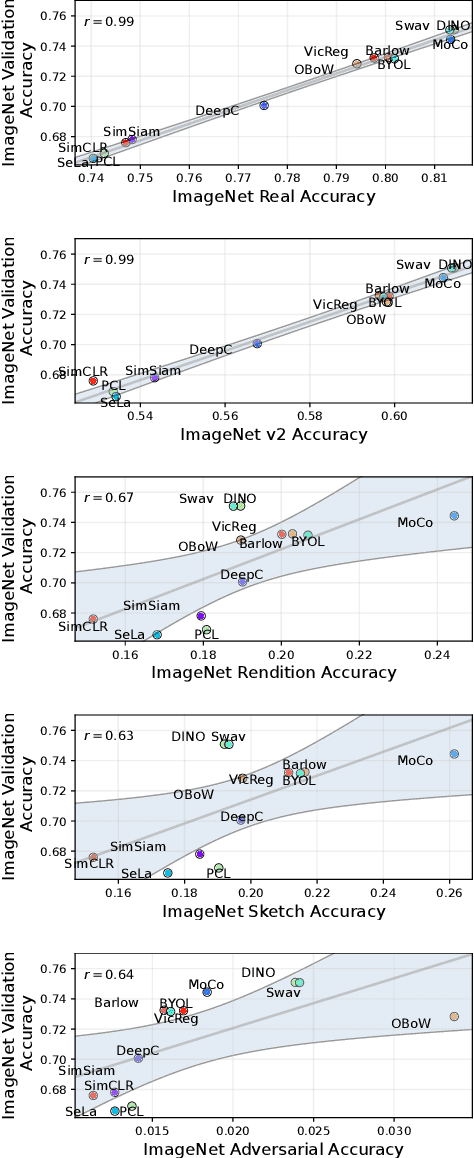
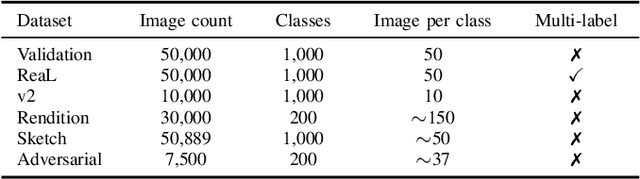
Abstract:Machine learning (ML) research strongly relies on benchmarks in order to determine the relative effectiveness of newly proposed models. Recently, a number of prominent research effort argued that a number of models that improve the state-of-the-art by a small margin tend to do so by winning what they call a "benchmark lottery". An important benchmark in the field of machine learning and computer vision is the ImageNet where newly proposed models are often showcased based on their performance on this dataset. Given the large number of self-supervised learning (SSL) frameworks that has been proposed in the past couple of years each coming with marginal improvements on the ImageNet dataset, in this work, we evaluate whether those marginal improvements on ImageNet translate to improvements on similar datasets or not. To do so, we investigate twelve popular SSL frameworks on five ImageNet variants and discover that models that seem to perform well on ImageNet may experience significant performance declines on similar datasets. Specifically, state-of-the-art frameworks such as DINO and Swav, which are praised for their performance, exhibit substantial drops in performance while MoCo and Barlow Twins displays comparatively good results. As a result, we argue that otherwise good and desirable properties of models remain hidden when benchmarking is only performed on the ImageNet validation set, making us call for more adequate benchmarking. To avoid the "benchmark lottery" on ImageNet and to ensure a fair benchmarking process, we investigate the usage of a unified metric that takes into account the performance of models on other ImageNet variant datasets.
Utilizing Mutations to Evaluate Interpretability of Neural Networks on Genomic Data
Dec 12, 2022
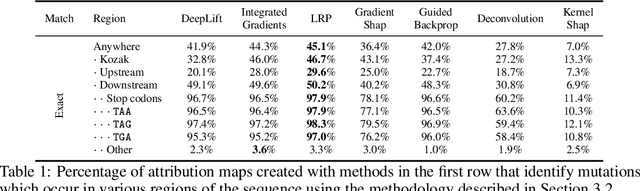
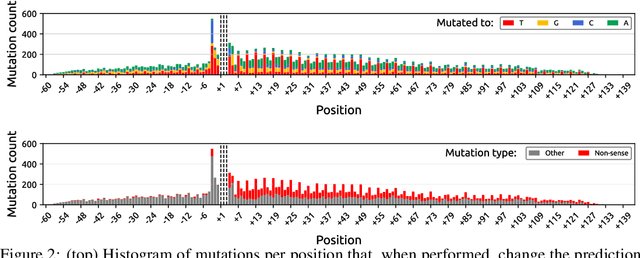

Abstract:Even though deep neural networks (DNNs) achieve state-of-the-art results for a number of problems involving genomic data, getting DNNs to explain their decision-making process has been a major challenge due to their black-box nature. One way to get DNNs to explain their reasoning for prediction is via attribution methods which are assumed to highlight the parts of the input that contribute to the prediction the most. Given the existence of numerous attribution methods and a lack of quantitative results on the fidelity of those methods, selection of an attribution method for sequence-based tasks has been mostly done qualitatively. In this work, we take a step towards identifying the most faithful attribution method by proposing a computational approach that utilizes point mutations. Providing quantitative results on seven popular attribution methods, we find Layerwise Relevance Propagation (LRP) to be the most appropriate one for translation initiation, with LRP identifying two important biological features for translation: the integrity of Kozak sequence as well as the detrimental effects of premature stop codons.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge