Siyang Leng
Utilizing Causal Network Markers to Identify Tipping Points ahead of Critical Transition
Dec 19, 2024



Abstract:Early-warning signals of delicate design are always used to predict critical transitions in complex systems, which makes it possible to render the systems far away from the catastrophic state by introducing timely interventions. Traditional signals including the dynamical network biomarker (DNB), based on statistical properties such as variance and autocorrelation of nodal dynamics, overlook directional interactions and thus have limitations in capturing underlying mechanisms and simultaneously sustaining robustness against noise perturbations. This paper therefore introduces a framework of causal network markers (CNMs) by incorporating causality indicators, which reflect the directional influence between variables. Actually, to detect and identify the tipping points ahead of critical transition, two markers are designed: CNM-GC for linear causality and CNM-TE for non-linear causality, as well as a functional representation of different causality indicators and a clustering technique to verify the system's dominant group. Through demonstrations using benchmark models and real-world datasets of epileptic seizure, the framework of CNMs shows higher predictive power and accuracy than the traditional DNB indicator. It is believed that, due to the versatility and scalability, the CNMs are suitable for comprehensively evaluating the systems. The most possible direction for application includes the identification of tipping points in clinical disease.
Deciphering interventional dynamical causality from non-intervention systems
Jun 29, 2024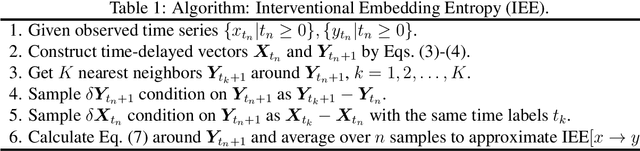
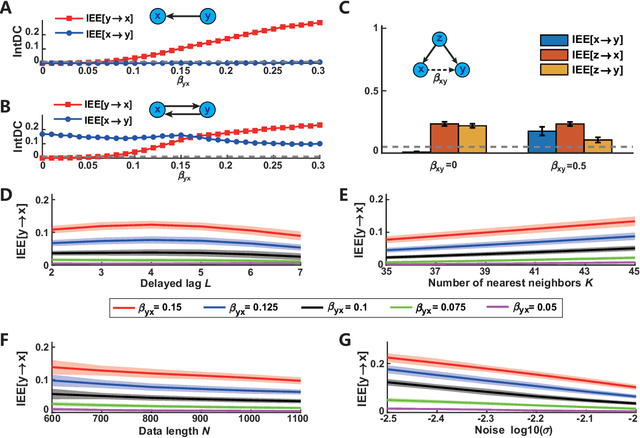
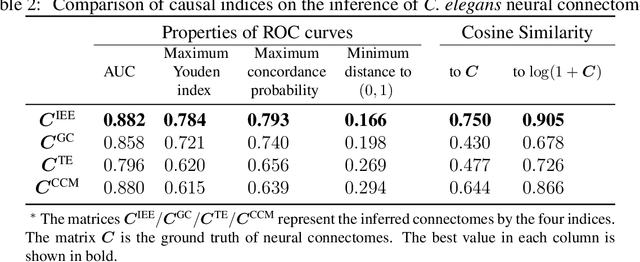
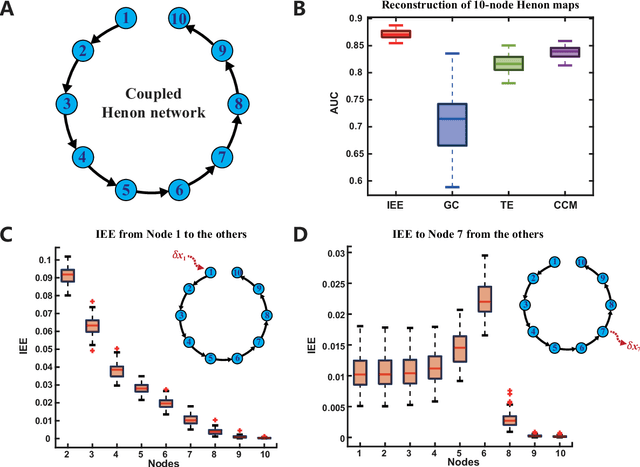
Abstract:Detecting and quantifying causality is a focal topic in the fields of science, engineering, and interdisciplinary studies. However, causal studies on non-intervention systems attract much attention but remain extremely challenging. To address this challenge, we propose a framework named Interventional Dynamical Causality (IntDC) for such non-intervention systems, along with its computational criterion, Interventional Embedding Entropy (IEE), to quantify causality. The IEE criterion theoretically and numerically enables the deciphering of IntDC solely from observational (non-interventional) time-series data, without requiring any knowledge of dynamical models or real interventions in the considered system. Demonstrations of performance showed the accuracy and robustness of IEE on benchmark simulated systems as well as real-world systems, including the neural connectomes of C. elegans, COVID-19 transmission networks in Japan, and regulatory networks surrounding key circadian genes.
UniG-Encoder: A Universal Feature Encoder for Graph and Hypergraph Node Classification
Aug 03, 2023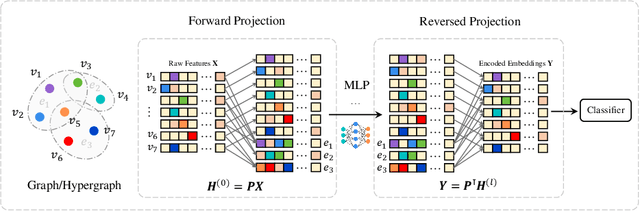
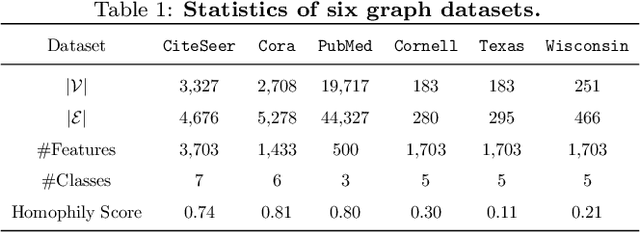
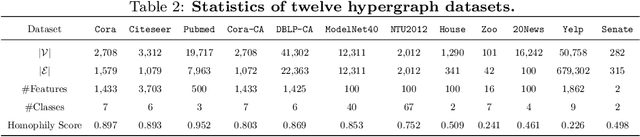
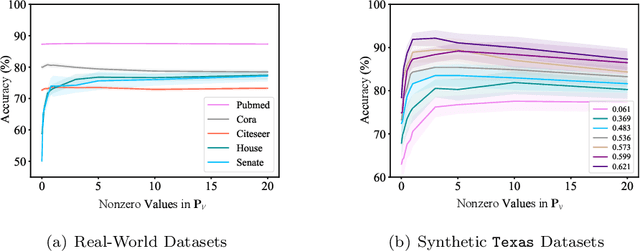
Abstract:Graph and hypergraph representation learning has attracted increasing attention from various research fields. Despite the decent performance and fruitful applications of Graph Neural Networks (GNNs), Hypergraph Neural Networks (HGNNs), and their well-designed variants, on some commonly used benchmark graphs and hypergraphs, they are outperformed by even a simple Multi-Layer Perceptron. This observation motivates a reexamination of the design paradigm of the current GNNs and HGNNs and poses challenges of extracting graph features effectively. In this work, a universal feature encoder for both graph and hypergraph representation learning is designed, called UniG-Encoder. The architecture starts with a forward transformation of the topological relationships of connected nodes into edge or hyperedge features via a normalized projection matrix. The resulting edge/hyperedge features, together with the original node features, are fed into a neural network. The encoded node embeddings are then derived from the reversed transformation, described by the transpose of the projection matrix, of the network's output, which can be further used for tasks such as node classification. The proposed architecture, in contrast to the traditional spectral-based and/or message passing approaches, simultaneously and comprehensively exploits the node features and graph/hypergraph topologies in an efficient and unified manner, covering both heterophilic and homophilic graphs. The designed projection matrix, encoding the graph features, is intuitive and interpretable. Extensive experiments are conducted and demonstrate the superior performance of the proposed framework on twelve representative hypergraph datasets and six real-world graph datasets, compared to the state-of-the-art methods. Our implementation is available online at https://github.com/MinhZou/UniG-Encoder.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge