Sharmila Upadhyaya
GSAP-ERE: Fine-Grained Scholarly Entity and Relation Extraction Focused on Machine Learning
Nov 12, 2025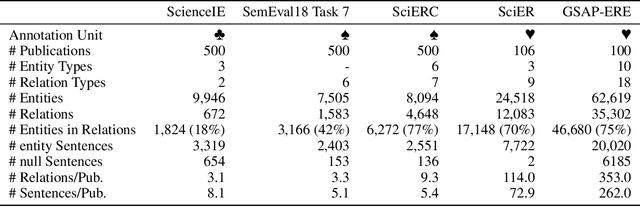
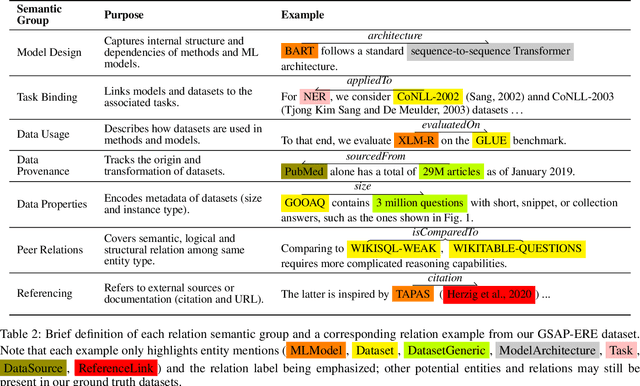
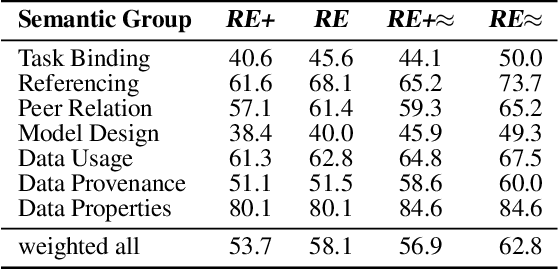
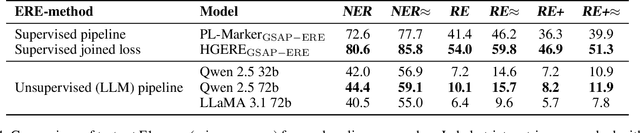
Abstract:Research in Machine Learning (ML) and AI evolves rapidly. Information Extraction (IE) from scientific publications enables to identify information about research concepts and resources on a large scale and therefore is a pathway to improve understanding and reproducibility of ML-related research. To extract and connect fine-grained information in ML-related research, e.g. method training and data usage, we introduce GSAP-ERE. It is a manually curated fine-grained dataset with 10 entity types and 18 semantically categorized relation types, containing mentions of 63K entities and 35K relations from the full text of 100 ML publications. We show that our dataset enables fine-tuned models to automatically extract information relevant for downstream tasks ranging from knowledge graph (KG) construction, to monitoring the computational reproducibility of AI research at scale. Additionally, we use our dataset as a test suite to explore prompting strategies for IE using Large Language Models (LLM). We observe that the performance of state-of-the-art LLM prompting methods is largely outperformed by our best fine-tuned baseline model (NER: 80.6%, RE: 54.0% for the fine-tuned model vs. NER: 44.4%, RE: 10.1% for the LLM). This disparity of performance between supervised models and unsupervised usage of LLMs suggests datasets like GSAP-ERE are needed to advance research in the domain of scholarly information extraction.
NFDI4DS Shared Tasks for Scholarly Document Processing
Sep 26, 2025Abstract:Shared tasks are powerful tools for advancing research through community-based standardised evaluation. As such, they play a key role in promoting findable, accessible, interoperable, and reusable (FAIR), as well as transparent and reproducible research practices. This paper presents an updated overview of twelve shared tasks developed and hosted under the German National Research Data Infrastructure for Data Science and Artificial Intelligence (NFDI4DS) consortium, covering a diverse set of challenges in scholarly document processing. Hosted at leading venues, the tasks foster methodological innovations and contribute open-access datasets, models, and tools for the broader research community, which are integrated into the consortium's research data infrastructure.
Utilizing Large Language Models for Named Entity Recognition in Traditional Chinese Medicine against COVID-19 Literature: Comparative Study
Aug 24, 2024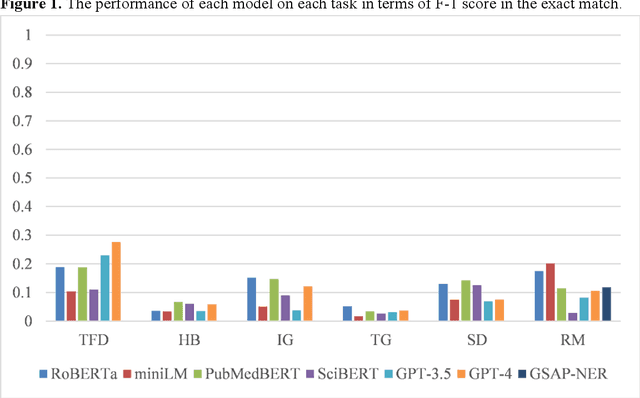
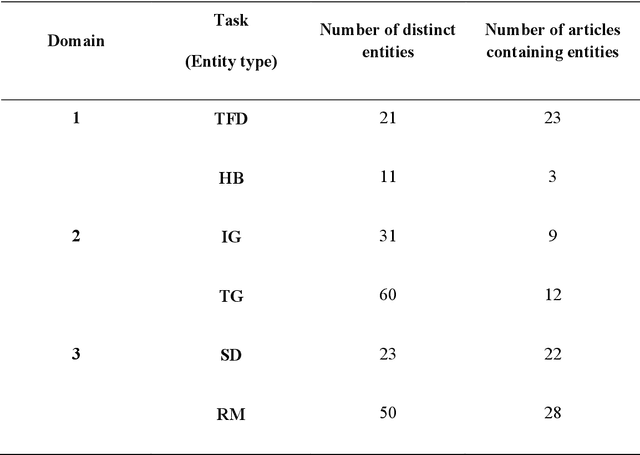
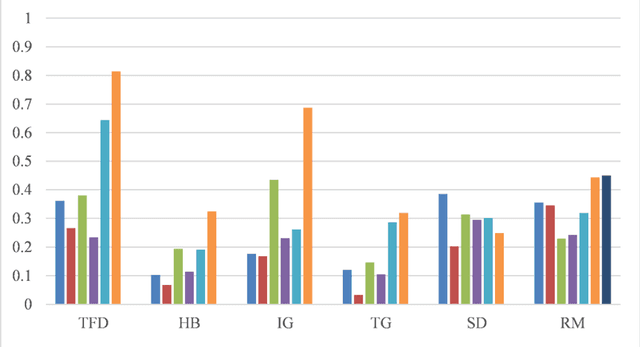
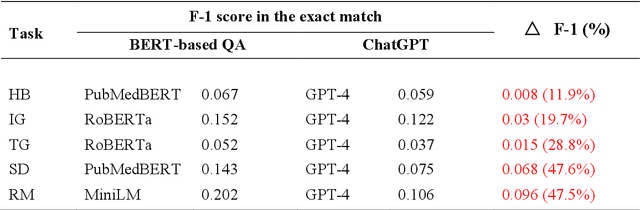
Abstract:Objective: To explore and compare the performance of ChatGPT and other state-of-the-art LLMs on domain-specific NER tasks covering different entity types and domains in TCM against COVID-19 literature. Methods: We established a dataset of 389 articles on TCM against COVID-19, and manually annotated 48 of them with 6 types of entities belonging to 3 domains as the ground truth, against which the NER performance of LLMs can be assessed. We then performed NER tasks for the 6 entity types using ChatGPT (GPT-3.5 and GPT-4) and 4 state-of-the-art BERT-based question-answering (QA) models (RoBERTa, MiniLM, PubMedBERT and SciBERT) without prior training on the specific task. A domain fine-tuned model (GSAP-NER) was also applied for a comprehensive comparison. Results: The overall performance of LLMs varied significantly in exact match and fuzzy match. In the fuzzy match, ChatGPT surpassed BERT-based QA models in 5 out of 6 tasks, while in exact match, BERT-based QA models outperformed ChatGPT in 5 out of 6 tasks but with a smaller F-1 difference. GPT-4 showed a significant advantage over other models in fuzzy match, especially on the entity type of TCM formula and the Chinese patent drug (TFD) and ingredient (IG). Although GPT-4 outperformed BERT-based models on entity type of herb, target, and research method, none of the F-1 scores exceeded 0.5. GSAP-NER, outperformed GPT-4 in terms of F-1 by a slight margin on RM. ChatGPT achieved considerably higher recalls than precisions, particularly in the fuzzy match. Conclusions: The NER performance of LLMs is highly dependent on the entity type, and their performance varies across application scenarios. ChatGPT could be a good choice for scenarios where high recall is favored. However, for knowledge acquisition in rigorous scenarios, neither ChatGPT nor BERT-based QA models are off-the-shelf tools for professional practitioners.
Enhancing Software Related Information Extraction with Generative Language Models through Single-Choice Question Answering
Apr 08, 2024Abstract:This paper describes our participation in the Shared Task on Software Mentions Disambiguation (SOMD), with a focus on improving relation extraction in scholarly texts through Generative Language Models (GLMs) using single-choice question-answering. The methodology prioritises the use of in-context learning capabilities of GLMs to extract software-related entities and their descriptive attributes, such as distributive information. Our approach uses Retrieval-Augmented Generation (RAG) techniques and GLMs for Named Entity Recognition (NER) and Attributive NER to identify relationships between extracted software entities, providing a structured solution for analysing software citations in academic literature. The paper provides a detailed description of our approach, demonstrating how using GLMs in a single-choice QA paradigm can greatly enhance IE methodologies. Our participation in the SOMD shared task highlights the importance of precise software citation practices and showcases our system's ability to overcome the challenges of disambiguating and extracting relationships between software mentions. This sets the groundwork for future research and development in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge