Shannon M. Lynch
STREAMLINE: An Automated Machine Learning Pipeline for Biomedicine Applied to Examine the Utility of Photography-Based Phenotypes for OSA Prediction Across International Sleep Centers
Dec 09, 2023



Abstract:While machine learning (ML) includes a valuable array of tools for analyzing biomedical data, significant time and expertise is required to assemble effective, rigorous, and unbiased pipelines. Automated ML (AutoML) tools seek to facilitate ML application by automating a subset of analysis pipeline elements. In this study we develop and validate a Simple, Transparent, End-to-end Automated Machine Learning Pipeline (STREAMLINE) and apply it to investigate the added utility of photography-based phenotypes for predicting obstructive sleep apnea (OSA); a common and underdiagnosed condition associated with a variety of health, economic, and safety consequences. STREAMLINE is designed to tackle biomedical binary classification tasks while adhering to best practices and accommodating complexity, scalability, reproducibility, customization, and model interpretation. Benchmarking analyses validated the efficacy of STREAMLINE across data simulations with increasingly complex patterns of association. Then we applied STREAMLINE to evaluate the utility of demographics (DEM), self-reported comorbidities (DX), symptoms (SYM), and photography-based craniofacial (CF) and intraoral (IO) anatomy measures in predicting any OSA or moderate/severe OSA using 3,111 participants from Sleep Apnea Global Interdisciplinary Consortium (SAGIC). OSA analyses identified a significant increase in ROC-AUC when adding CF to DEM+DX+SYM to predict moderate/severe OSA. A consistent but non-significant increase in PRC-AUC was observed with the addition of each subsequent feature set to predict any OSA, with CF and IO yielding minimal improvements. Application of STREAMLINE to OSA data suggests that CF features provide additional value in predicting moderate/severe OSA, but neither CF nor IO features meaningfully improved the prediction of any OSA beyond established demographics, comorbidity and symptom characteristics.
LCS-DIVE: An Automated Rule-based Machine Learning Visualization Pipeline for Characterizing Complex Associations in Classification
Apr 26, 2021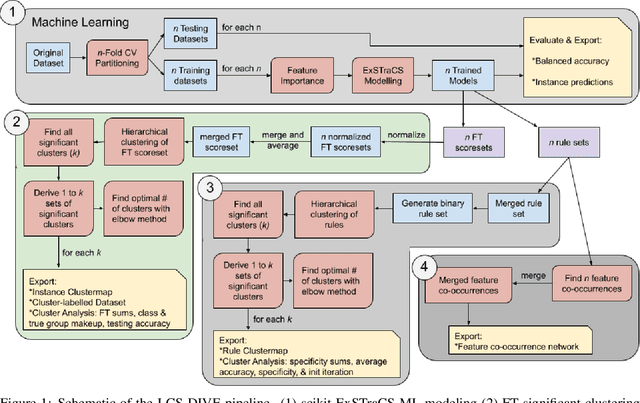
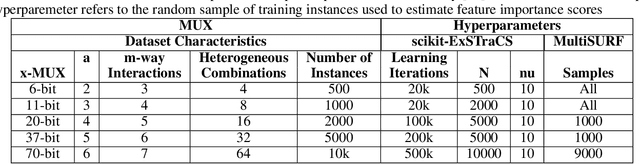
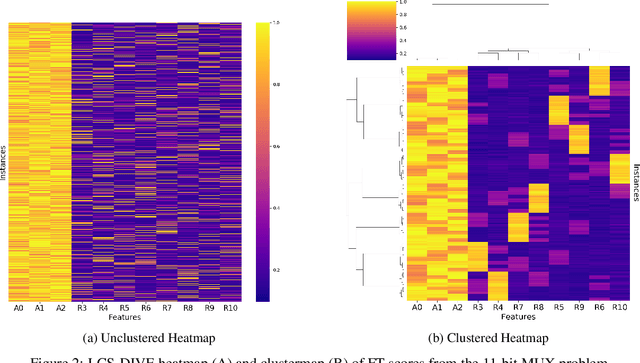
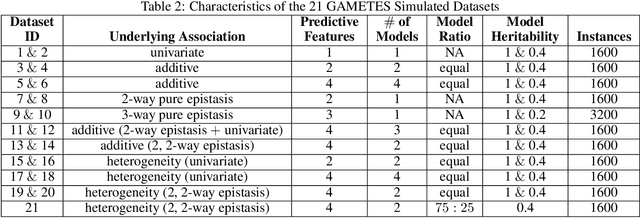
Abstract:Machine learning (ML) research has yielded powerful tools for training accurate prediction models despite complex multivariate associations (e.g. interactions and heterogeneity). In fields such as medicine, improved interpretability of ML modeling is required for knowledge discovery, accountability, and fairness. Rule-based ML approaches such as Learning Classifier Systems (LCSs) strike a balance between predictive performance and interpretability in complex, noisy domains. This work introduces the LCS Discovery and Visualization Environment (LCS-DIVE), an automated LCS model interpretation pipeline for complex biomedical classification. LCS-DIVE conducts modeling using a new scikit-learn implementation of ExSTraCS, an LCS designed to overcome noise and scalability in biomedical data mining yielding human readable IF:THEN rules as well as feature-tracking scores for each training sample. LCS-DIVE leverages feature-tracking scores and/or rules to automatically guide characterization of (1) feature importance (2) underlying additive, epistatic, and/or heterogeneous patterns of association, and (3) model-driven heterogeneous instance subgroups via clustering, visualization generation, and cluster interrogation. LCS-DIVE was evaluated over a diverse set of simulated genetic and benchmark datasets encoding a variety of complex multivariate associations, demonstrating its ability to differentiate between them and then applied to characterize associations within a real-world study of pancreatic cancer.
A Rigorous Machine Learning Analysis Pipeline for Biomedical Binary Classification: Application in Pancreatic Cancer Nested Case-control Studies with Implications for Bias Assessments
Sep 08, 2020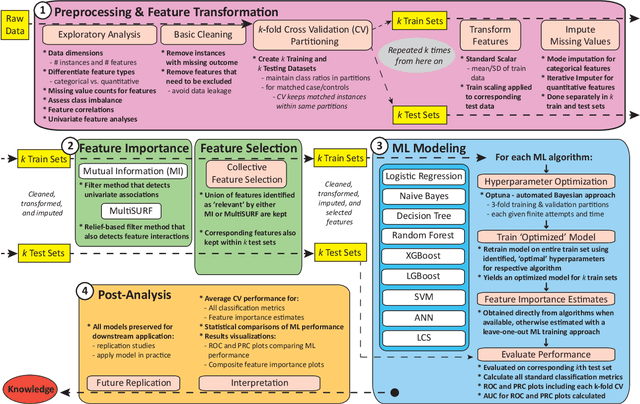
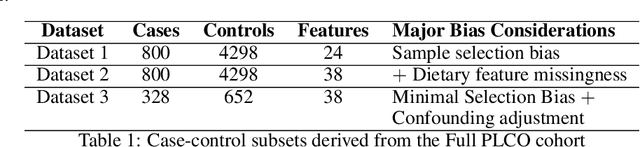
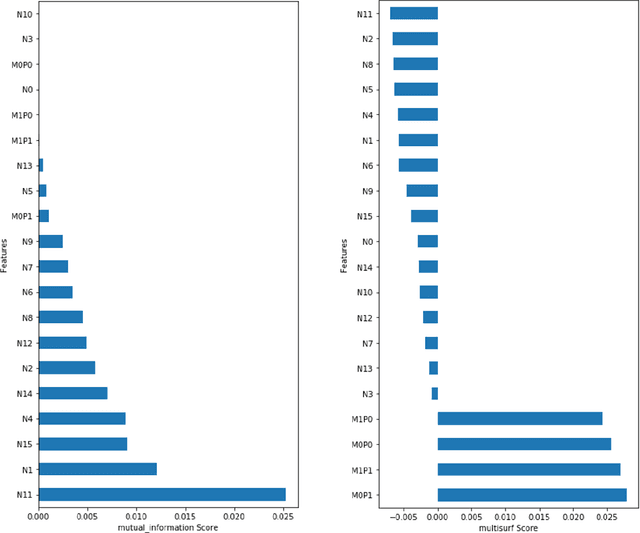
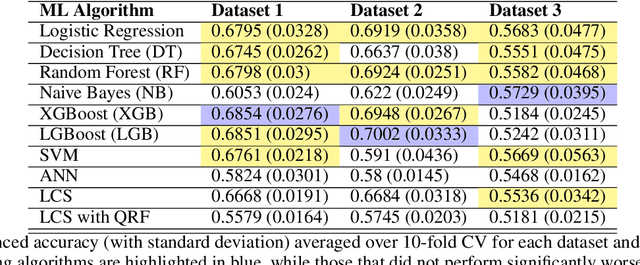
Abstract:Machine learning (ML) offers a collection of powerful approaches for detecting and modeling associations, often applied to data having a large number of features and/or complex associations. Currently, there are many tools to facilitate implementing custom ML analyses (e.g. scikit-learn). Interest is also increasing in automated ML packages, which can make it easier for non-experts to apply ML and have the potential to improve model performance. ML permeates most subfields of biomedical research with varying levels of rigor and correct usage. Tremendous opportunities offered by ML are frequently offset by the challenge of assembling comprehensive analysis pipelines, and the ease of ML misuse. In this work we have laid out and assembled a complete, rigorous ML analysis pipeline focused on binary classification (i.e. case/control prediction), and applied this pipeline to both simulated and real world data. At a high level, this 'automated' but customizable pipeline includes a) exploratory analysis, b) data cleaning and transformation, c) feature selection, d) model training with 9 established ML algorithms, each with hyperparameter optimization, and e) thorough evaluation, including appropriate metrics, statistical analyses, and novel visualizations. This pipeline organizes the many subtle complexities of ML pipeline assembly to illustrate best practices to avoid bias and ensure reproducibility. Additionally, this pipeline is the first to compare established ML algorithms to 'ExSTraCS', a rule-based ML algorithm with the unique capability of interpretably modeling heterogeneous patterns of association. While designed to be widely applicable we apply this pipeline to an epidemiological investigation of established and newly identified risk factors for pancreatic cancer to evaluate how different sources of bias might be handled by ML algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge