Pranshu Suri
STREAMLINE: A Simple, Transparent, End-To-End Automated Machine Learning Pipeline Facilitating Data Analysis and Algorithm Comparison
Jun 23, 2022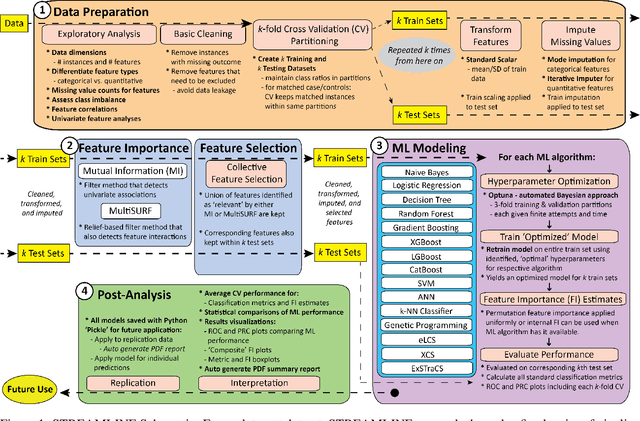
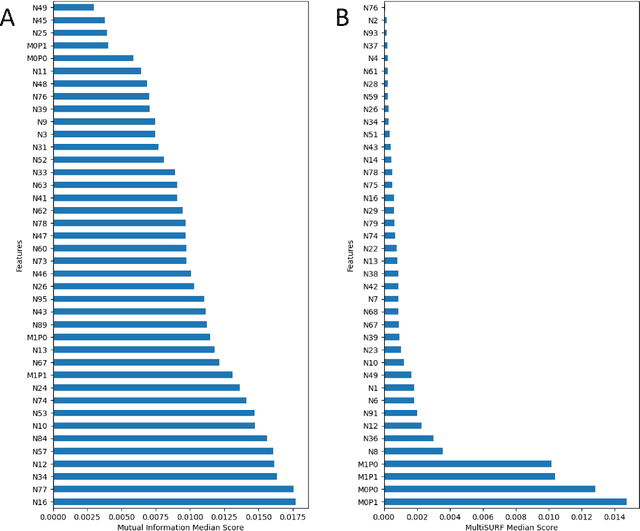
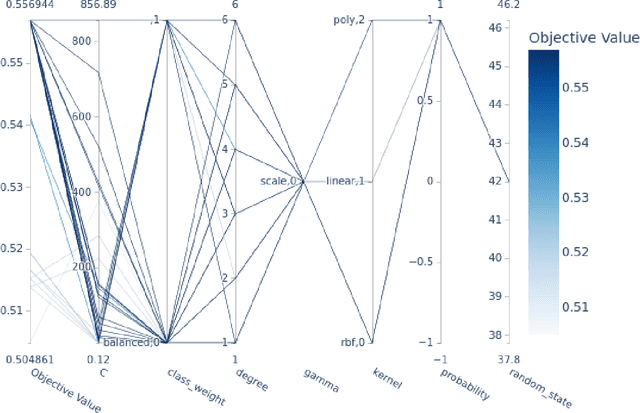
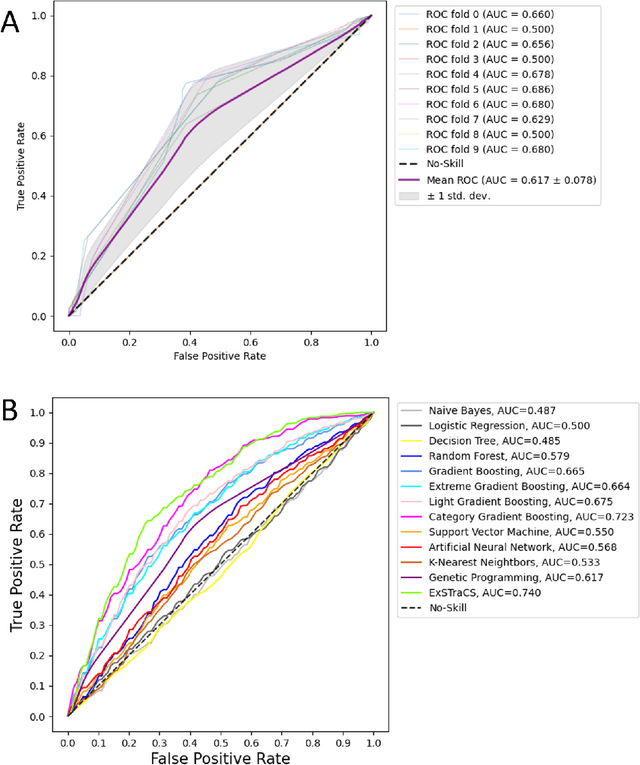
Abstract:Machine learning (ML) offers powerful methods for detecting and modeling associations often in data with large feature spaces and complex associations. Many useful tools/packages (e.g. scikit-learn) have been developed to make the various elements of data handling, processing, modeling, and interpretation accessible. However, it is not trivial for most investigators to assemble these elements into a rigorous, replicatable, unbiased, and effective data analysis pipeline. Automated machine learning (AutoML) seeks to address these issues by simplifying the process of ML analysis for all. Here, we introduce STREAMLINE, a simple, transparent, end-to-end AutoML pipeline designed as a framework to easily conduct rigorous ML modeling and analysis (limited initially to binary classification). STREAMLINE is specifically designed to compare performance between datasets, ML algorithms, and other AutoML tools. It is unique among other autoML tools by offering a fully transparent and consistent baseline of comparison using a carefully designed series of pipeline elements including: (1) exploratory analysis, (2) basic data cleaning, (3) cross validation partitioning, (4) data scaling and imputation, (5) filter-based feature importance estimation, (6) collective feature selection, (7) ML modeling with `Optuna' hyperparameter optimization across 15 established algorithms (including less well-known Genetic Programming and rule-based ML), (8) evaluation across 16 classification metrics, (9) model feature importance estimation, (10) statistical significance comparisons, and (11) automatically exporting all results, plots, a PDF summary report, and models that can be easily applied to replication data.
A Rigorous Machine Learning Analysis Pipeline for Biomedical Binary Classification: Application in Pancreatic Cancer Nested Case-control Studies with Implications for Bias Assessments
Sep 08, 2020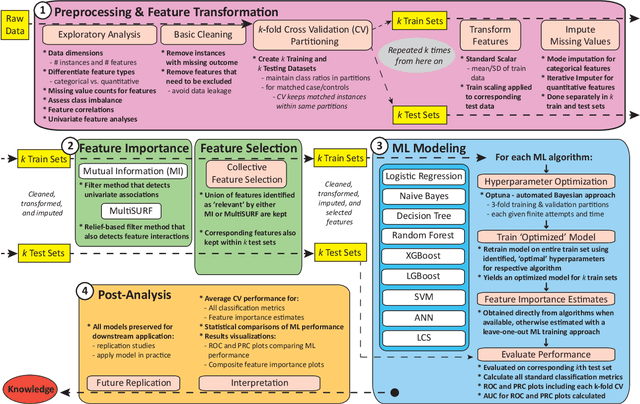
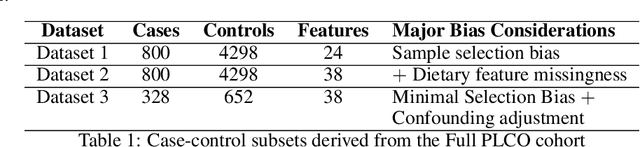
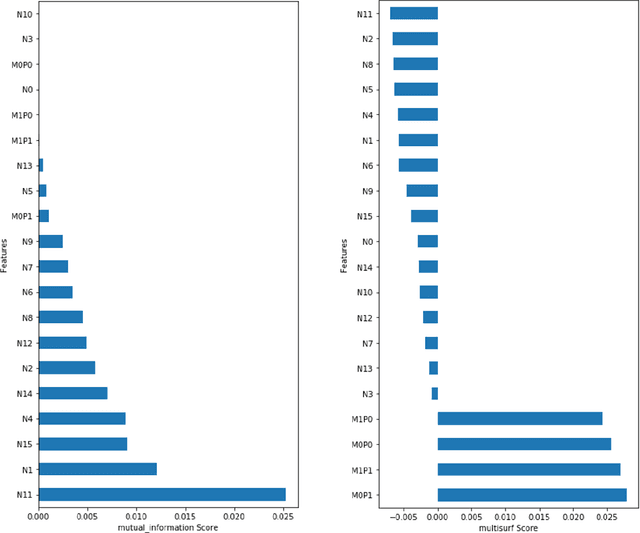
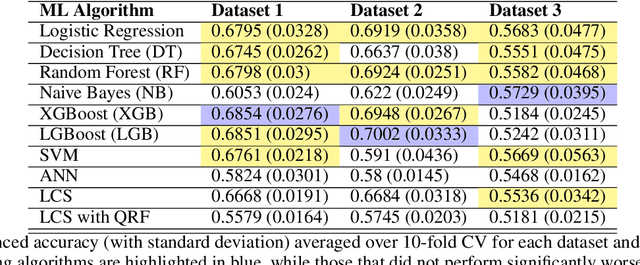
Abstract:Machine learning (ML) offers a collection of powerful approaches for detecting and modeling associations, often applied to data having a large number of features and/or complex associations. Currently, there are many tools to facilitate implementing custom ML analyses (e.g. scikit-learn). Interest is also increasing in automated ML packages, which can make it easier for non-experts to apply ML and have the potential to improve model performance. ML permeates most subfields of biomedical research with varying levels of rigor and correct usage. Tremendous opportunities offered by ML are frequently offset by the challenge of assembling comprehensive analysis pipelines, and the ease of ML misuse. In this work we have laid out and assembled a complete, rigorous ML analysis pipeline focused on binary classification (i.e. case/control prediction), and applied this pipeline to both simulated and real world data. At a high level, this 'automated' but customizable pipeline includes a) exploratory analysis, b) data cleaning and transformation, c) feature selection, d) model training with 9 established ML algorithms, each with hyperparameter optimization, and e) thorough evaluation, including appropriate metrics, statistical analyses, and novel visualizations. This pipeline organizes the many subtle complexities of ML pipeline assembly to illustrate best practices to avoid bias and ensure reproducibility. Additionally, this pipeline is the first to compare established ML algorithms to 'ExSTraCS', a rule-based ML algorithm with the unique capability of interpretably modeling heterogeneous patterns of association. While designed to be widely applicable we apply this pipeline to an epidemiological investigation of established and newly identified risk factors for pancreatic cancer to evaluate how different sources of bias might be handled by ML algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge