Shahina Kunhimon
Hierarchical Self-Supervised Adversarial Training for Robust Vision Models in Histopathology
Mar 13, 2025Abstract:Adversarial attacks pose significant challenges for vision models in critical fields like healthcare, where reliability is essential. Although adversarial training has been well studied in natural images, its application to biomedical and microscopy data remains limited. Existing self-supervised adversarial training methods overlook the hierarchical structure of histopathology images, where patient-slide-patch relationships provide valuable discriminative signals. To address this, we propose Hierarchical Self-Supervised Adversarial Training (HSAT), which exploits these properties to craft adversarial examples using multi-level contrastive learning and integrate it into adversarial training for enhanced robustness. We evaluate HSAT on multiclass histopathology dataset OpenSRH and the results show that HSAT outperforms existing methods from both biomedical and natural image domains. HSAT enhances robustness, achieving an average gain of 54.31% in the white-box setting and reducing performance drops to 3-4% in the black-box setting, compared to 25-30% for the baseline. These results set a new benchmark for adversarial training in this domain, paving the way for more robust models. Our Code for training and evaluation is available at https://github.com/HashmatShadab/HSAT.
UniMed-CLIP: Towards a Unified Image-Text Pretraining Paradigm for Diverse Medical Imaging Modalities
Dec 13, 2024



Abstract:Vision-Language Models (VLMs) trained via contrastive learning have achieved notable success in natural image tasks. However, their application in the medical domain remains limited due to the scarcity of openly accessible, large-scale medical image-text datasets. Existing medical VLMs either train on closed-source proprietary or relatively small open-source datasets that do not generalize well. Similarly, most models remain specific to a single or limited number of medical imaging domains, again restricting their applicability to other modalities. To address this gap, we introduce UniMed, a large-scale, open-source multi-modal medical dataset comprising over 5.3 million image-text pairs across six diverse imaging modalities: X-ray, CT, MRI, Ultrasound, Pathology, and Fundus. UniMed is developed using a data-collection framework that leverages Large Language Models (LLMs) to transform modality-specific classification datasets into image-text formats while incorporating existing image-text data from the medical domain, facilitating scalable VLM pretraining. Using UniMed, we trained UniMed-CLIP, a unified VLM for six modalities that significantly outperforms existing generalist VLMs and matches modality-specific medical VLMs, achieving notable gains in zero-shot evaluations. For instance, UniMed-CLIP improves over BiomedCLIP (trained on proprietary data) by an absolute gain of +12.61, averaged over 21 datasets, while using 3x less training data. To facilitate future research, we release UniMed dataset, training codes, and models at https://github.com/mbzuai-oryx/UniMed-CLIP.
Language Guided Domain Generalized Medical Image Segmentation
Apr 03, 2024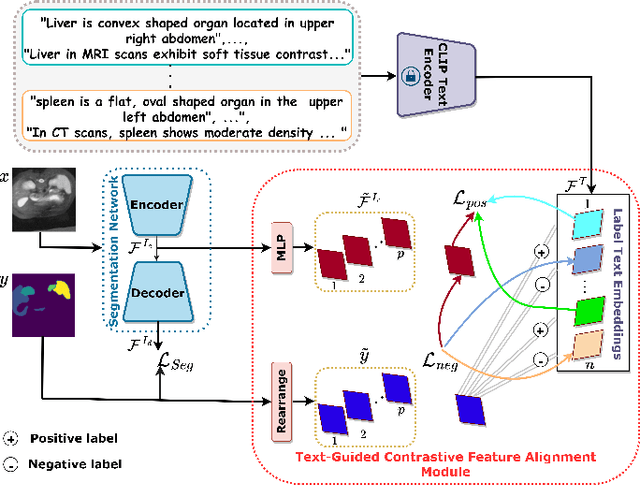
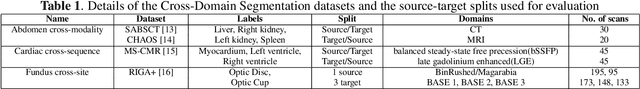
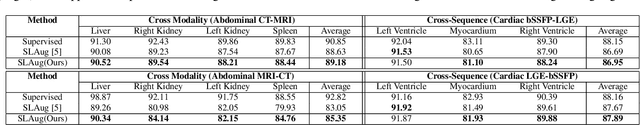

Abstract:Single source domain generalization (SDG) holds promise for more reliable and consistent image segmentation across real-world clinical settings particularly in the medical domain, where data privacy and acquisition cost constraints often limit the availability of diverse datasets. Depending solely on visual features hampers the model's capacity to adapt effectively to various domains, primarily because of the presence of spurious correlations and domain-specific characteristics embedded within the image features. Incorporating text features alongside visual features is a potential solution to enhance the model's understanding of the data, as it goes beyond pixel-level information to provide valuable context. Textual cues describing the anatomical structures, their appearances, and variations across various imaging modalities can guide the model in domain adaptation, ultimately contributing to more robust and consistent segmentation. In this paper, we propose an approach that explicitly leverages textual information by incorporating a contrastive learning mechanism guided by the text encoder features to learn a more robust feature representation. We assess the effectiveness of our text-guided contrastive feature alignment technique in various scenarios, including cross-modality, cross-sequence, and cross-site settings for different segmentation tasks. Our approach achieves favorable performance against existing methods in literature. Our code and model weights are available at https://github.com/ShahinaKK/LG_SDG.git.
Learnable Weight Initialization for Volumetric Medical Image Segmentation
Jun 28, 2023



Abstract:Hybrid volumetric medical image segmentation models, combining the advantages of local convolution and global attention, have recently received considerable attention. While mainly focusing on architectural modifications, most existing hybrid approaches still use conventional data-independent weight initialization schemes which restrict their performance due to ignoring the inherent volumetric nature of the medical data. To address this issue, we propose a learnable weight initialization approach that utilizes the available medical training data to effectively learn the contextual and structural cues via the proposed self-supervised objectives. Our approach is easy to integrate into any hybrid model and requires no external training data. Experiments on multi-organ and lung cancer segmentation tasks demonstrate the effectiveness of our approach, leading to state-of-the-art segmentation performance. Our proposed data-dependent initialization approach performs favorably as compared to the Swin-UNETR model pretrained using large-scale datasets on multi-organ segmentation task. Our source code and models are available at: https://github.com/ShahinaKK/LWI-VMS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge