Samuli Junttila
Multispectral airborne laser scanning for tree species classification: a benchmark of machine learning and deep learning algorithms
Apr 19, 2025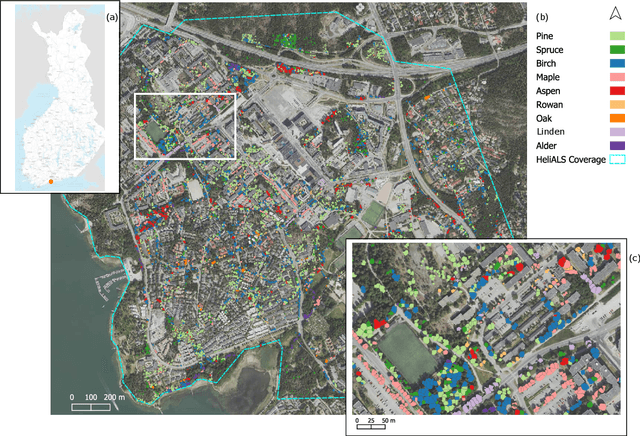
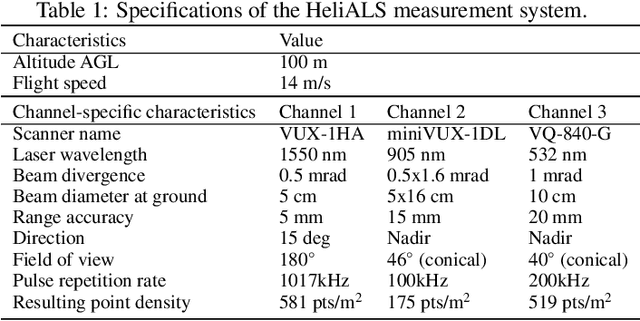

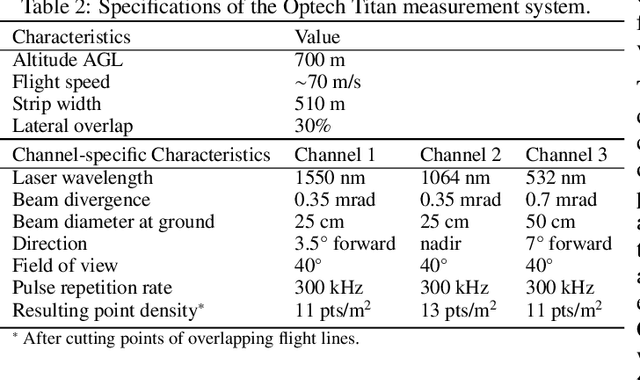
Abstract:Climate-smart and biodiversity-preserving forestry demands precise information on forest resources, extending to the individual tree level. Multispectral airborne laser scanning (ALS) has shown promise in automated point cloud processing and tree segmentation, but challenges remain in identifying rare tree species and leveraging deep learning techniques. This study addresses these gaps by conducting a comprehensive benchmark of machine learning and deep learning methods for tree species classification. For the study, we collected high-density multispectral ALS data (>1000 pts/m$^2$) at three wavelengths using the FGI-developed HeliALS system, complemented by existing Optech Titan data (35 pts/m$^2$), to evaluate the species classification accuracy of various algorithms in a test site located in Southern Finland. Based on 5261 test segments, our findings demonstrate that point-based deep learning methods, particularly a point transformer model, outperformed traditional machine learning and image-based deep learning approaches on high-density multispectral point clouds. For the high-density ALS dataset, a point transformer model provided the best performance reaching an overall (macro-average) accuracy of 87.9% (74.5%) with a training set of 1065 segments and 92.0% (85.1%) with 5000 training segments. The best image-based deep learning method, DetailView, reached an overall (macro-average) accuracy of 84.3% (63.9%), whereas a random forest (RF) classifier achieved an overall (macro-average) accuracy of 83.2% (61.3%). Importantly, the overall classification accuracy of the point transformer model on the HeliALS data increased from 73.0% with no spectral information to 84.7% with single-channel reflectance, and to 87.9% with spectral information of all the three channels.
ADA-Net: Attention-Guided Domain Adaptation Network with Contrastive Learning for Standing Dead Tree Segmentation Using Aerial Imagery
Apr 05, 2025Abstract:Information on standing dead trees is important for understanding forest ecosystem functioning and resilience but has been lacking over large geographic regions. Climate change has caused large-scale tree mortality events that can remain undetected due to limited data. In this study, we propose a novel method for segmenting standing dead trees using aerial multispectral orthoimages. Because access to annotated datasets has been a significant problem in forest remote sensing due to the need for forest expertise, we introduce a method for domain transfer by leveraging domain adaptation to learn a transformation from a source domain X to target domain Y. In this Image-to-Image translation task, we aim to utilize available annotations in the target domain by pre-training a segmentation network. When images from a new study site without annotations are introduced (source domain X), these images are transformed into the target domain. Then, transfer learning is applied by inferring the pre-trained network on domain-adapted images. In addition to investigating the feasibility of current domain adaptation approaches for this objective, we propose a novel approach called the Attention-guided Domain Adaptation Network (ADA-Net) with enhanced contrastive learning. Accordingly, the ADA-Net approach provides new state-of-the-art domain adaptation performance levels outperforming existing approaches. We have evaluated the proposed approach using two datasets from Finland and the US. The USA images are converted to the Finland domain, and we show that the synthetic USA2Finland dataset exhibits similar characteristics to the Finland domain images. The software implementation is shared at https://github.com/meteahishali/ADA-Net. The data is publicly available at https://www.kaggle.com/datasets/meteahishali/aerial-imagery-for-standing-dead-tree-segmentation.
Dual-Task Learning for Dead Tree Detection and Segmentation with Hybrid Self-Attention U-Nets in Aerial Imagery
Mar 27, 2025Abstract:Mapping standing dead trees is critical for assessing forest health, monitoring biodiversity, and mitigating wildfire risks, for which aerial imagery has proven useful. However, dense canopy structures, spectral overlaps between living and dead vegetation, and over-segmentation errors limit the reliability of existing methods. This study introduces a hybrid postprocessing framework that refines deep learning-based tree segmentation by integrating watershed algorithms with adaptive filtering, enhancing boundary delineation, and reducing false positives in complex forest environments. Tested on high-resolution aerial imagery from boreal forests, the framework improved instance-level segmentation accuracy by 41.5% and reduced positional errors by 57%, demonstrating robust performance in densely vegetated regions. By balancing detection accuracy and over-segmentation artifacts, the method enabled the precise identification of individual dead trees, which is critical for ecological monitoring. The framework's computational efficiency supports scalable applications, such as wall-to-wall tree mortality mapping over large geographic regions using aerial or satellite imagery. These capabilities directly benefit wildfire risk assessment (identifying fuel accumulations), carbon stock estimation (tracking emissions from decaying biomass), and precision forestry (targeting salvage loggings). By bridging advanced remote sensing techniques with practical forest management needs, this work advances tools for large-scale ecological conservation and climate resilience planning.
Benchmarking tree species classification from proximally-sensed laser scanning data: introducing the FOR-species20K dataset
Aug 12, 2024Abstract:Proximally-sensed laser scanning offers significant potential for automated forest data capture, but challenges remain in automatically identifying tree species without additional ground data. Deep learning (DL) shows promise for automation, yet progress is slowed by the lack of large, diverse, openly available labeled datasets of single tree point clouds. This has impacted the robustness of DL models and the ability to establish best practices for species classification. To overcome these challenges, the FOR-species20K benchmark dataset was created, comprising over 20,000 tree point clouds from 33 species, captured using terrestrial (TLS), mobile (MLS), and drone laser scanning (ULS) across various European forests, with some data from other regions. This dataset enables the benchmarking of DL models for tree species classification, including both point cloud-based (PointNet++, MinkNet, MLP-Mixer, DGCNNs) and multi-view image-based methods (SimpleView, DetailView, YOLOv5). 2D image-based models generally performed better (average OA = 0.77) than 3D point cloud-based models (average OA = 0.72), with consistent results across different scanning platforms and sensors. The top model, DetailView, was particularly robust, handling data imbalances well and generalizing effectively across tree sizes. The FOR-species20K dataset, available at https://zenodo.org/records/13255198, is a key resource for developing and benchmarking DL models for tree species classification using laser scanning data, providing a foundation for future advancements in the field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge