Saimunur Rahman
Learning Compact Channel Correlation Representation for LiDAR Place Recognition
Sep 24, 2024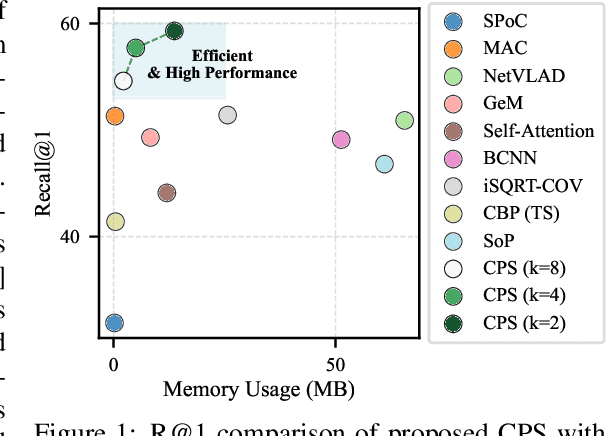
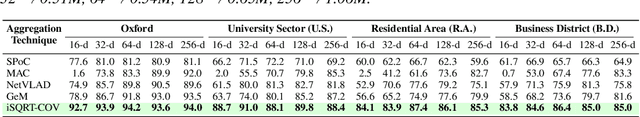
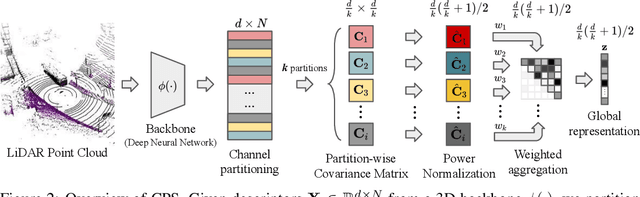
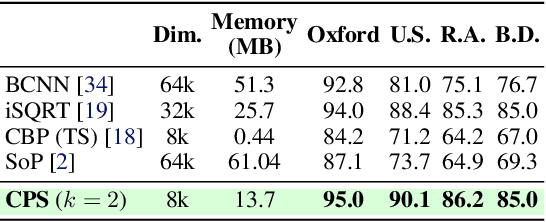
Abstract:This paper presents a novel approach to learn compact channel correlation representation for LiDAR place recognition, called C3R, aimed at reducing the computational burden and dimensionality associated with traditional covariance pooling methods for place recognition tasks. Our method partitions the feature matrix into smaller groups, computes group-wise covariance matrices, and aggregates them via a learnable aggregation strategy. Matrix power normalization is applied to ensure stability. Theoretical analyses are also given to demonstrate the effectiveness of the proposed method, including its ability to preserve permutation invariance and maintain high mutual information between the original features and the aggregated representation. We conduct extensive experiments on four large-scale, public LiDAR place recognition datasets including Oxford RobotCar, In-house, MulRan, and WildPlaces datasets to validate our approach's superiority in accuracy, and robustness. Furthermore, we provide the quantitative results of our approach for a deeper understanding. The code will be released upon acceptance.
PseudoNeg-MAE: Self-Supervised Point Cloud Learning using Conditional Pseudo-Negative Embeddings
Sep 24, 2024Abstract:We propose PseudoNeg-MAE, a novel self-supervised learning framework that enhances global feature representation of point cloud mask autoencoder by making them both discriminative and sensitive to transformations. Traditional contrastive learning methods focus on achieving invariance, which can lead to the loss of valuable transformation-related information. In contrast, PseudoNeg-MAE explicitly models the relationship between original and transformed data points using a parametric network COPE, which learns the localized displacements caused by transformations within the latent space. However, jointly training COPE with the MAE leads to undesirable trivial solutions where COPE outputs collapse to an identity. To address this, we introduce a novel loss function incorporating pseudo-negatives, which effectively penalizes these trivial invariant solutions and promotes transformation sensitivity in the embeddings. We validate PseudoNeg-MAE on shape classification and relative pose estimation tasks, where PseudoNeg-MAE achieves state-of-the-art performance on the ModelNet40 and ScanObjectNN datasets under challenging evaluation protocols and demonstrates superior accuracy in estimating relative poses. These results show the effectiveness of PseudoNeg-MAE in learning discriminative and transformation-sensitive representations.
Learning Partial Correlation based Deep Visual Representation for Image Classification
Apr 26, 2023



Abstract:Visual representation based on covariance matrix has demonstrates its efficacy for image classification by characterising the pairwise correlation of different channels in convolutional feature maps. However, pairwise correlation will become misleading once there is another channel correlating with both channels of interest, resulting in the ``confounding'' effect. For this case, ``partial correlation'' which removes the confounding effect shall be estimated instead. Nevertheless, reliably estimating partial correlation requires to solve a symmetric positive definite matrix optimisation, known as sparse inverse covariance estimation (SICE). How to incorporate this process into CNN remains an open issue. In this work, we formulate SICE as a novel structured layer of CNN. To ensure end-to-end trainability, we develop an iterative method to solve the above matrix optimisation during forward and backward propagation steps. Our work obtains a partial correlation based deep visual representation and mitigates the small sample problem often encountered by covariance matrix estimation in CNN. Computationally, our model can be effectively trained with GPU and works well with a large number of channels of advanced CNNs. Experiments show the efficacy and superior classification performance of our deep visual representation compared to covariance matrix based counterparts.
Deep Learning based HEp-2 Image Classification: A Comprehensive Review
Nov 20, 2019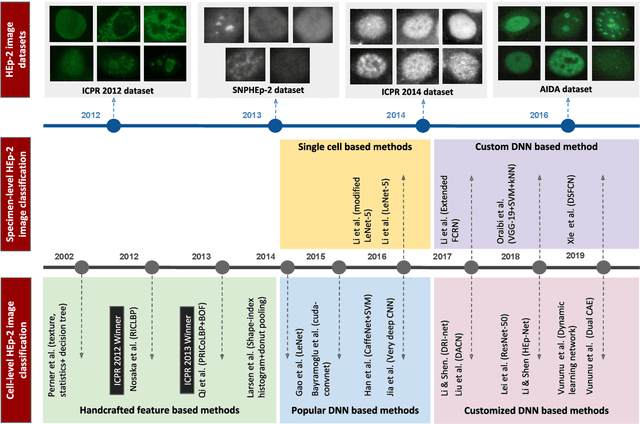
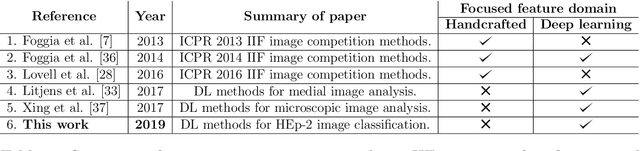
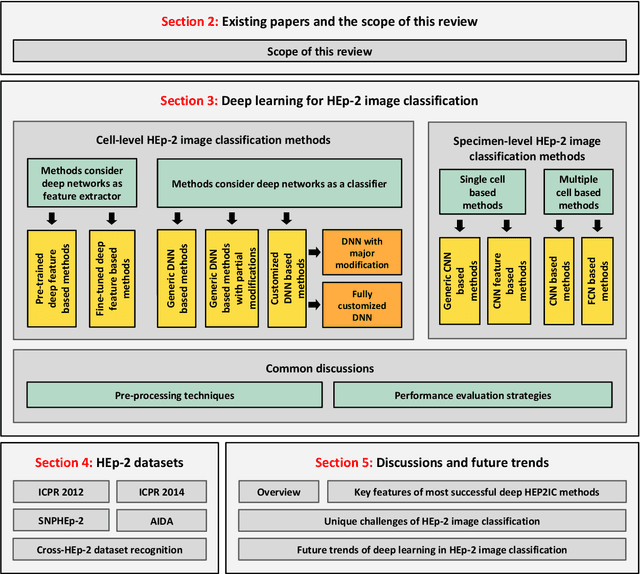
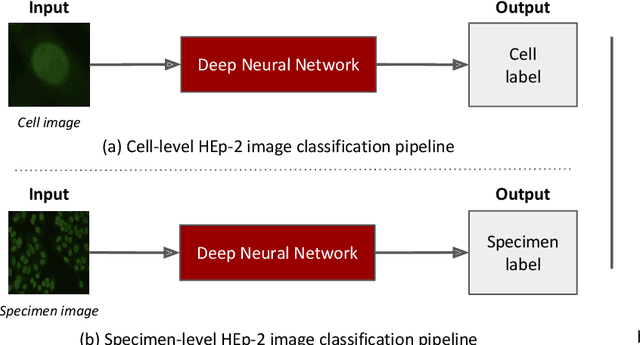
Abstract:Classification of HEp-2 cell patterns plays a significant role in the indirect immunofluorescence test for identifying autoimmune diseases in the human body. Many automatic HEp-2 cell classification methods have been proposed in recent years, amongst which deep learning based methods have shown impressive performance. This paper provides a comprehensive review of the existing deep learning based HEp-2 cell image classification methods. These methods perform HEp-2 image classification in two levels, namely, cell-level and specimen-level. Both levels are covered in this review. In each level, the methods are organized with a deep network usage based taxonomy. The core idea, notable achievements, and key advantages and weakness of each method are critically analyzed. Furthermore, a concise review of the existing HEp-2 datasets that are commonly used in the literature is given. The paper ends with an overview of the current state-of-the-arts and a discussion on novel opportunities and future research directions in this field. It is hoped that this paper would give readers a comprehensive reference of this novel, challenging, and thriving field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge