Saga Helgadottir
Roadmap on Deep Learning for Microscopy
Mar 07, 2023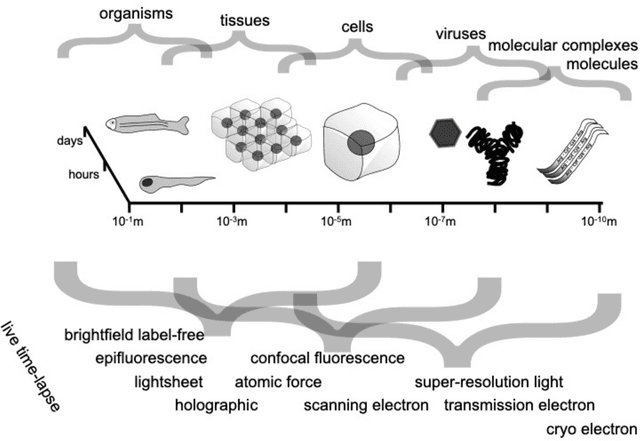
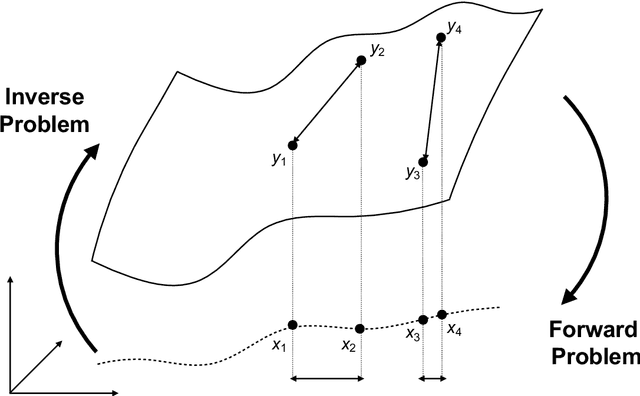
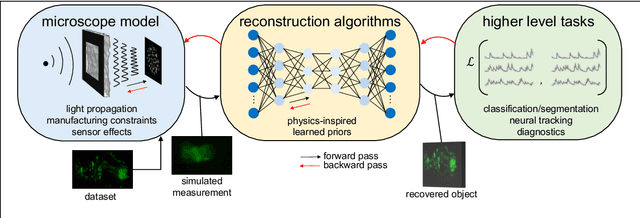
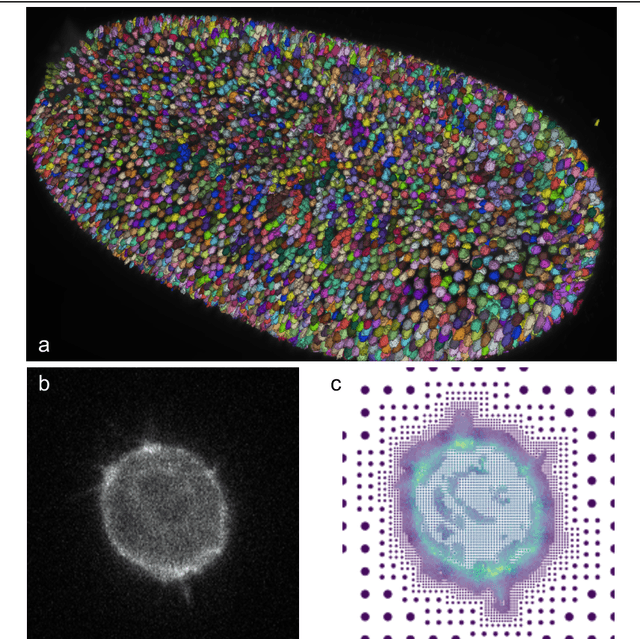
Abstract:Through digital imaging, microscopy has evolved from primarily being a means for visual observation of life at the micro- and nano-scale, to a quantitative tool with ever-increasing resolution and throughput. Artificial intelligence, deep neural networks, and machine learning are all niche terms describing computational methods that have gained a pivotal role in microscopy-based research over the past decade. This Roadmap is written collectively by prominent researchers and encompasses selected aspects of how machine learning is applied to microscopy image data, with the aim of gaining scientific knowledge by improved image quality, automated detection, segmentation, classification and tracking of objects, and efficient merging of information from multiple imaging modalities. We aim to give the reader an overview of the key developments and an understanding of possibilities and limitations of machine learning for microscopy. It will be of interest to a wide cross-disciplinary audience in the physical sciences and life sciences.
Extracting quantitative biological information from brightfield cell images using deep learning
Dec 23, 2020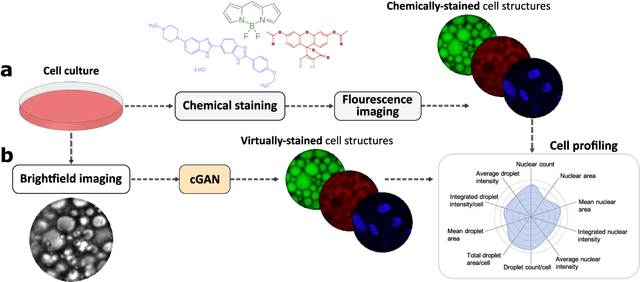
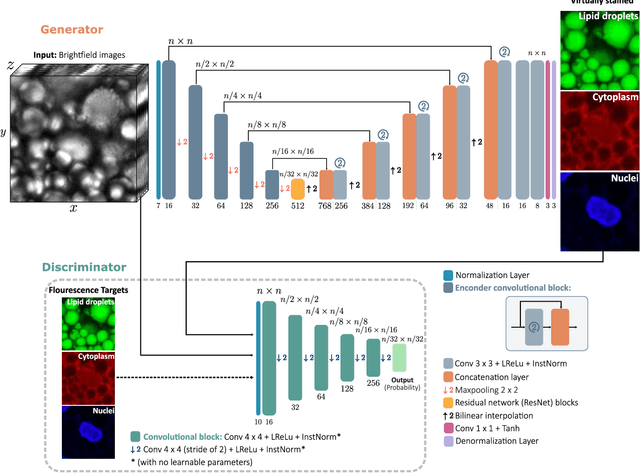
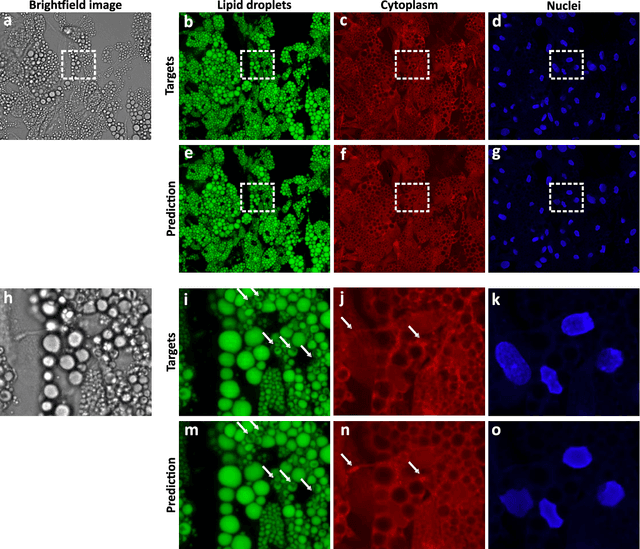
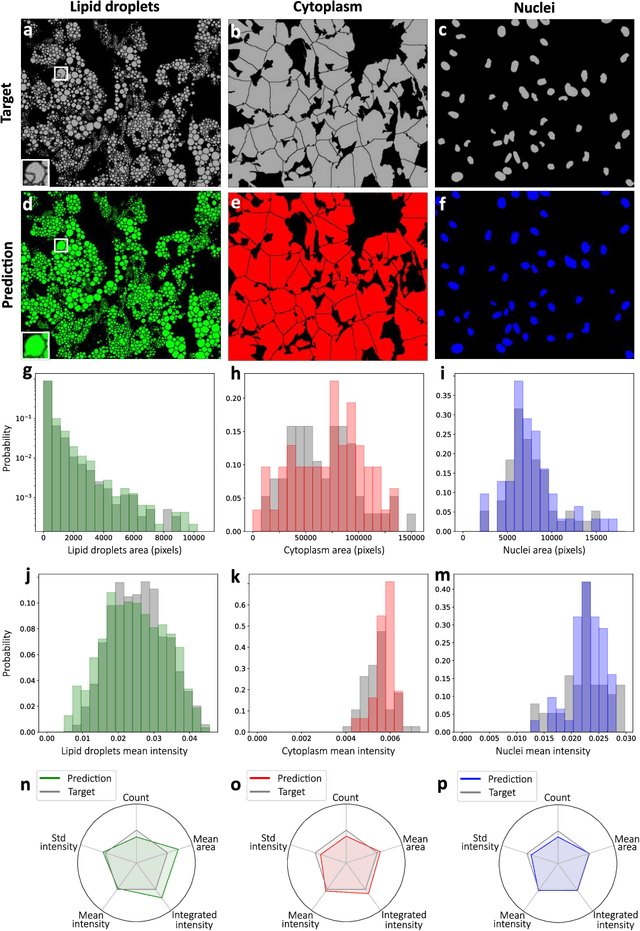
Abstract:Quantitative analysis of cell structures is essential for biomedical and pharmaceutical research. The standard imaging approach relies on fluorescence microscopy, where cell structures of interest are labeled by chemical staining techniques. However, these techniques are often invasive and sometimes even toxic to the cells, in addition to being time-consuming, labor-intensive, and expensive. Here, we introduce an alternative deep-learning-powered approach based on the analysis of brightfield images by a conditional generative adversarial neural network (cGAN). We show that this approach can extract information from the brightfield images to generate virtually-stained images, which can be used in subsequent downstream quantitative analyses of cell structures. Specifically, we train a cGAN to virtually stain lipid droplets, cytoplasm, and nuclei using brightfield images of human stem-cell-derived fat cells (adipocytes), which are of particular interest for nanomedicine and vaccine development. Subsequently, we use these virtually-stained images to extract quantitative measures about these cell structures. Generating virtually-stained fluorescence images is less invasive, less expensive, and more reproducible than standard chemical staining; furthermore, it frees up the fluorescence microscopy channels for other analytical probes, thus increasing the amount of information that can be extracted from each cell.
Improving epidemic testing and containment strategies using machine learning
Nov 23, 2020



Abstract:Containment of epidemic outbreaks entails great societal and economic costs. Cost-effective containment strategies rely on efficiently identifying infected individuals, making the best possible use of the available testing resources. Therefore, quickly identifying the optimal testing strategy is of critical importance. Here, we demonstrate that machine learning can be used to identify which individuals are most beneficial to test, automatically and dynamically adapting the testing strategy to the characteristics of the disease outbreak. Specifically, we simulate an outbreak using the archetypal susceptible-infectious-recovered (SIR) model and we use data about the first confirmed cases to train a neural network that learns to make predictions about the rest of the population. Using these prediction, we manage to contain the outbreak more effectively and more quickly than with standard approaches. Furthermore, we demonstrate how this method can be used also when there is a possibility of reinfection (SIRS model) to efficiently eradicate an endemic disease.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge