Robert Hoehndorf
GRAFT: Decoupling Ranking and Calibration for Survival Analysis
Feb 08, 2026Abstract:Survival analysis is complicated by censored data, high-dimensional features, and non-linear interactions. Classical models are interpretable but restrictive, while deep learning models are flexible but often non-interpretable and sensitive to noise. We propose GRAFT (Gated Residual Accelerated Failure Time), a novel AFT model that decouples prognostic ranking from calibration. GRAFT's hybrid architecture combines a linear AFT model with a non-linear residual neural network, and it also integrates stochastic gates for automatic, end-to-end feature selection. The model is trained by directly optimizing a differentiable, C-index-aligned ranking loss using stochastic conditional imputation from local Kaplan-Meier estimators. In public benchmarks, GRAFT outperforms baselines in discrimination and calibration, while remaining robust and sparse in high-noise settings.
Fully Geometric Multi-Hop Reasoning on Knowledge Graphs with Transitive Relations
May 18, 2025Abstract:Geometric embedding methods have shown to be useful for multi-hop reasoning on knowledge graphs by mapping entities and logical operations to geometric regions and geometric transformations, respectively. Geometric embeddings provide direct interpretability framework for queries. However, current methods have only leveraged the geometric construction of entities, failing to map logical operations to geometric transformations and, instead, using neural components to learn these operations. We introduce GeometrE, a geometric embedding method for multi-hop reasoning, which does not require learning the logical operations and enables full geometric interpretability. Additionally, unlike previous methods, we introduce a transitive loss function and show that it can preserve the logical rule $\forall a,b,c: r(a,b) \land r(b,c) \to r(a,c)$. Our experiments show that GeometrE outperforms current state-of-the-art methods on standard benchmark datasets.
Causal knowledge graph analysis identifies adverse drug effects
May 11, 2025Abstract:Knowledge graphs and structural causal models have each proven valuable for organizing biomedical knowledge and estimating causal effects, but remain largely disconnected: knowledge graphs encode qualitative relationships focusing on facts and deductive reasoning without formal probabilistic semantics, while causal models lack integration with background knowledge in knowledge graphs and have no access to the deductive reasoning capabilities that knowledge graphs provide. To bridge this gap, we introduce a novel formulation of Causal Knowledge Graphs (CKGs) which extend knowledge graphs with formal causal semantics, preserving their deductive capabilities while enabling principled causal inference. CKGs support deconfounding via explicitly marked causal edges and facilitate hypothesis formulation aligned with both encoded and entailed background knowledge. We constructed a Drug-Disease CKG (DD-CKG) integrating disease progression pathways, drug indications, side-effects, and hierarchical disease classification to enable automated large-scale mediation analysis. Applied to UK Biobank and MIMIC-IV cohorts, we tested whether drugs mediate effects between indications and downstream disease progression, adjusting for confounders inferred from the DD-CKG. Our approach successfully reproduced known adverse drug reactions with high precision while identifying previously undocumented significant candidate adverse effects. Further validation through side effect similarity analysis demonstrated that combining our predicted drug effects with established databases significantly improves the prediction of shared drug indications, supporting the clinical relevance of our novel findings. These results demonstrate that our methodology provides a generalizable, knowledge-driven framework for scalable causal inference.
DELE: Deductive $\mathcal{EL}^{++} \thinspace $ Embeddings for Knowledge Base Completion
Nov 03, 2024



Abstract:Ontology embeddings map classes, relations, and individuals in ontologies into $\mathbb{R}^n$, and within $\mathbb{R}^n$ similarity between entities can be computed or new axioms inferred. For ontologies in the Description Logic $\mathcal{EL}^{++}$, several embedding methods have been developed that explicitly generate models of an ontology. However, these methods suffer from some limitations; they do not distinguish between statements that are unprovable and provably false, and therefore they may use entailed statements as negatives. Furthermore, they do not utilize the deductive closure of an ontology to identify statements that are inferred but not asserted. We evaluated a set of embedding methods for $\mathcal{EL}^{++}$ ontologies, incorporating several modifications that aim to make use of the ontology deductive closure. In particular, we designed novel negative losses that account both for the deductive closure and different types of negatives and formulated evaluation methods for knowledge base completion. We demonstrate that our embedding methods improve over the baseline ontology embedding in the task of knowledge base or ontology completion.
Large-Scale Knowledge Integration for Enhanced Molecular Property Prediction
Oct 15, 2024Abstract:Pre-training machine learning models on molecular properties has proven effective for generating robust and generalizable representations, which is critical for advancements in drug discovery and materials science. While recent work has primarily focused on data-driven approaches, the KANO model introduces a novel paradigm by incorporating knowledge-enhanced pre-training. In this work, we expand upon KANO by integrating the large-scale ChEBI knowledge graph, which includes 2,840 functional groups -- significantly more than the original 82 used in KANO. We explore two approaches, Replace and Integrate, to incorporate this extensive knowledge into the KANO framework. Our results demonstrate that including ChEBI leads to improved performance on 9 out of 14 molecular property prediction datasets. This highlights the importance of utilizing a larger and more diverse set of functional groups to enhance molecular representations for property predictions. Code: github.com/Yasir-Ghunaim/KANO-ChEBI
Ontology Embedding: A Survey of Methods, Applications and Resources
Jun 16, 2024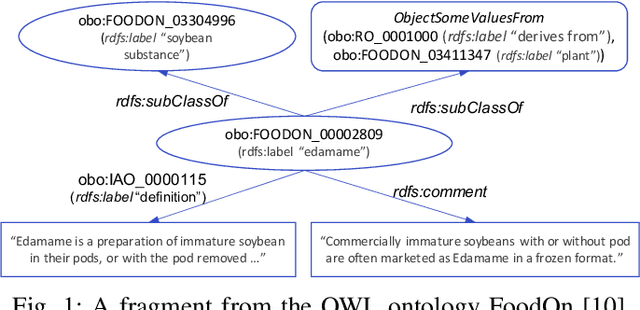
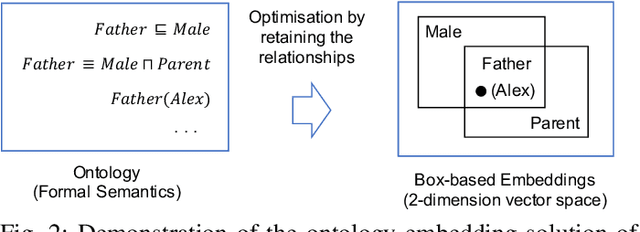
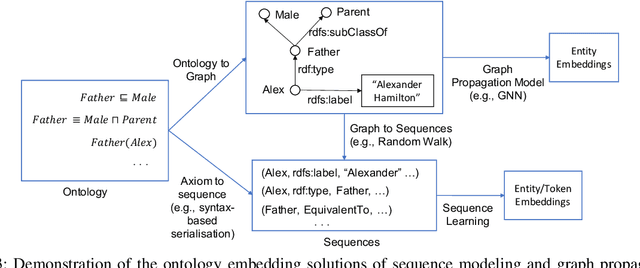
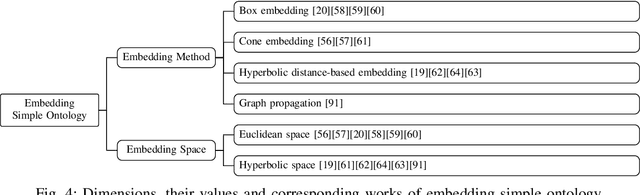
Abstract:Ontologies are widely used for representing domain knowledge and meta data, playing an increasingly important role in Information Systems, the Semantic Web, Bioinformatics and many other domains. However, logical reasoning that ontologies can directly support are quite limited in learning, approximation and prediction. One straightforward solution is to integrate statistical analysis and machine learning. To this end, automatically learning vector representation for knowledge of an ontology i.e., ontology embedding has been widely investigated in recent years. Numerous papers have been published on ontology embedding, but a lack of systematic reviews hinders researchers from gaining a comprehensive understanding of this field. To bridge this gap, we write this survey paper, which first introduces different kinds of semantics of ontologies, and formally defines ontology embedding from the perspectives of both mathematics and machine learning, as well as its property of faithfulness. Based on this, it systematically categorises and analyses a relatively complete set of over 80 papers, according to the ontologies and semantics that they aim at, and their technical solutions including geometric modeling, sequence modeling and graph propagation. This survey also introduces the applications of ontology embedding in ontology engineering, machine learning augmentation and life sciences, presents a new library mOWL, and discusses the challenges and future directions.
Enhancing Geometric Ontology Embeddings for $\mathcal{EL}^{++}$ with Negative Sampling and Deductive Closure Filtering
May 08, 2024



Abstract:Ontology embeddings map classes, relations, and individuals in ontologies into $\mathbb{R}^n$, and within $\mathbb{R}^n$ similarity between entities can be computed or new axioms inferred. For ontologies in the Description Logic $\mathcal{EL}^{++}$, several embedding methods have been developed that explicitly generate models of an ontology. However, these methods suffer from some limitations; they do not distinguish between statements that are unprovable and provably false, and therefore they may use entailed statements as negatives. Furthermore, they do not utilize the deductive closure of an ontology to identify statements that are inferred but not asserted. We evaluated a set of embedding methods for $\mathcal{EL}^{++}$ ontologies based on high-dimensional ball representation of concept descriptions, incorporating several modifications that aim to make use of the ontology deductive closure. In particular, we designed novel negative losses that account both for the deductive closure and different types of negatives. We demonstrate that our embedding methods improve over the baseline ontology embedding in the task of knowledge base or ontology completion.
Stylized Projected GAN: A Novel Architecture for Fast and Realistic Image Generation
Jul 30, 2023Abstract:Generative Adversarial Networks are used for generating the data using a generator and a discriminator, GANs usually produce high-quality images, but training GANs in an adversarial setting is a difficult task. GANs require high computation power and hyper-parameter regularization for converging. Projected GANs tackle the training difficulty of GANs by using transfer learning to project the generated and real samples into a pre-trained feature space. Projected GANs improve the training time and convergence but produce artifacts in the generated images which reduce the quality of the generated samples, we propose an optimized architecture called Stylized Projected GANs which integrates the mapping network of the Style GANs with Skip Layer Excitation of Fast GAN. The integrated modules are incorporated within the generator architecture of the Fast GAN to mitigate the problem of artifacts in the generated images.
An Open-Source Knowledge Graph Ecosystem for the Life Sciences
Jul 11, 2023
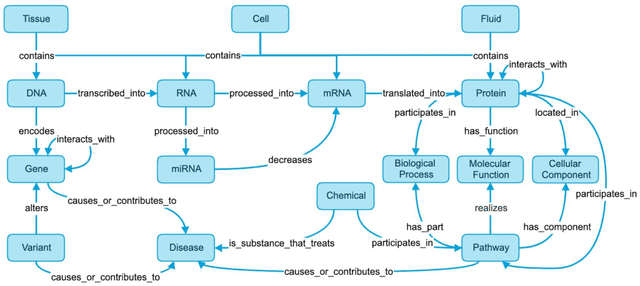
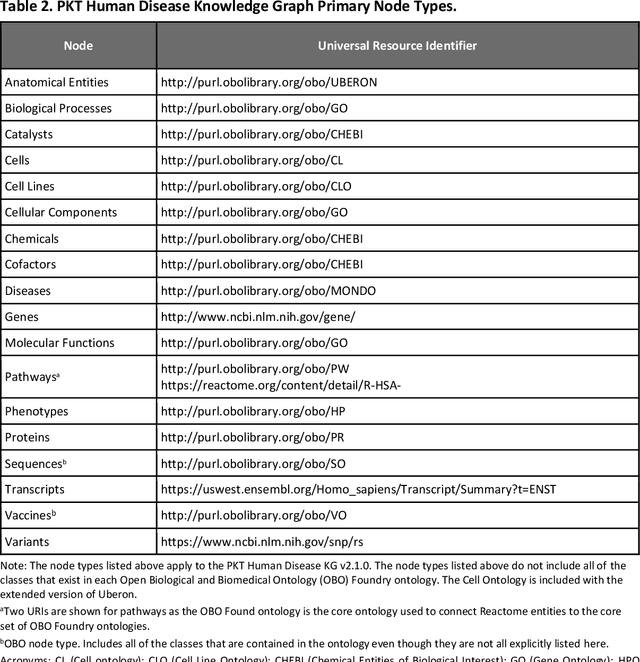
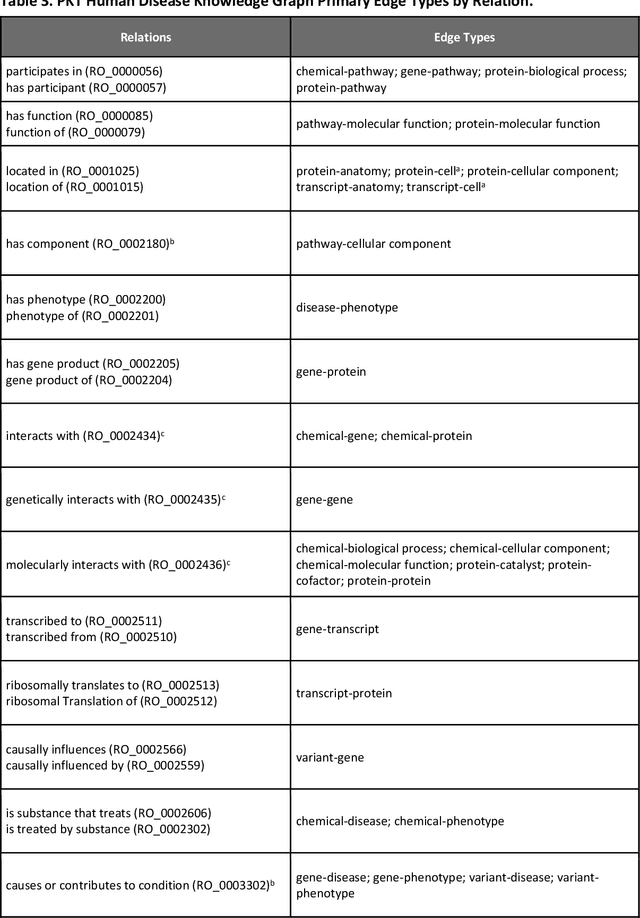
Abstract:Translational research requires data at multiple scales of biological organization. Advancements in sequencing and multi-omics technologies have increased the availability of these data but researchers face significant integration challenges. Knowledge graphs (KGs) are used to model complex phenomena, and methods exist to automatically construct them. However, tackling complex biomedical integration problems requires flexibility in the way knowledge is modeled. Moreover, existing KG construction methods provide robust tooling at the cost of fixed or limited choices among knowledge representation models. PheKnowLator (Phenotype Knowledge Translator) is a semantic ecosystem for automating the FAIR (Findable, Accessible, Interoperable, and Reusable) construction of ontologically grounded KGs with fully customizable knowledge representation. The ecosystem includes KG construction resources (e.g., data preparation APIs), analysis tools (e.g., SPARQL endpoints and abstraction algorithms), and benchmarks (e.g., prebuilt KGs and embeddings). We evaluate the ecosystem by surveying open-source KG construction methods and analyzing its computational performance when constructing 12 large-scale KGs. With flexible knowledge representation, PheKnowLator enables fully customizable KGs without compromising performance or usability.
CatE: Embedding $\mathcal{ALC}$ ontologies using category-theoretical semantics
May 11, 2023



Abstract:Machine learning with Semantic Web ontologies follows several strategies, one of which involves projecting ontologies into graph structures and applying graph embeddings or graph-based machine learning methods to the resulting graphs. Several methods have been developed that project ontology axioms into graphs. However, these methods are limited in the type of axioms they can project (totality), whether they are invertible (injectivity), and how they exploit semantic information. These limitations restrict the kind of tasks to which they can be applied. Category-theoretical semantics of logic languages formalizes interpretations using categories instead of sets, and categories have a graph-like structure. We developed CatE, which uses the category-theoretical formulation of the semantics of the Description Logic $\mathcal{ALC}$ to generate a graph representation for ontology axioms. The CatE projection is total and injective, and therefore overcomes limitations of other graph-based ontology embedding methods which are generally not invertible. We apply CatE to a number of different tasks, including deductive and inductive reasoning, and we demonstrate that CatE improves over state of the art ontology embedding methods. Furthermore, we show that CatE can also outperform model-theoretic ontology embedding methods in machine learning tasks in the biomedical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge