Rhiju Das
Predictive models of RNA degradation through dual crowdsourcing
Oct 14, 2021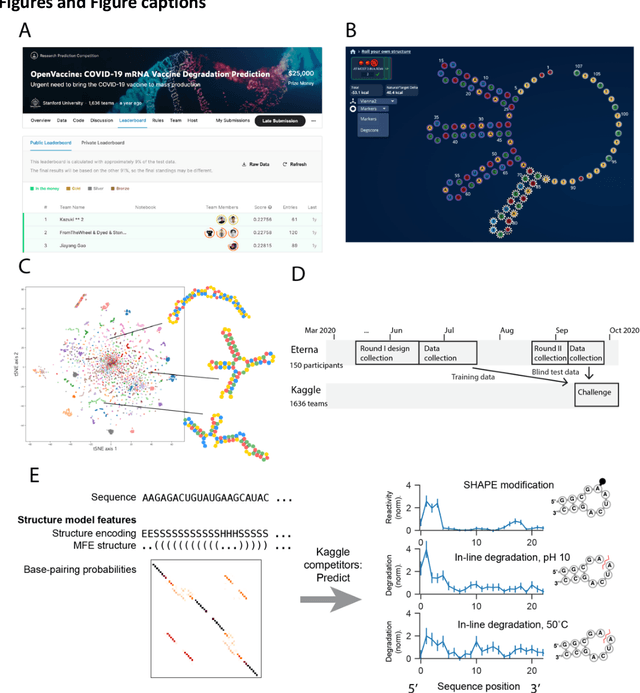
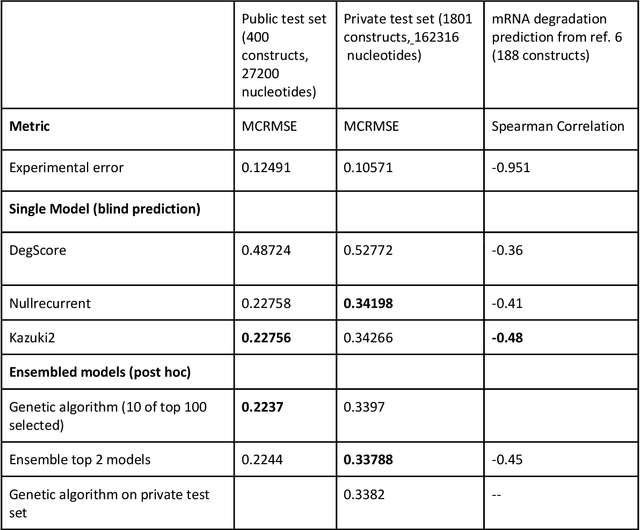
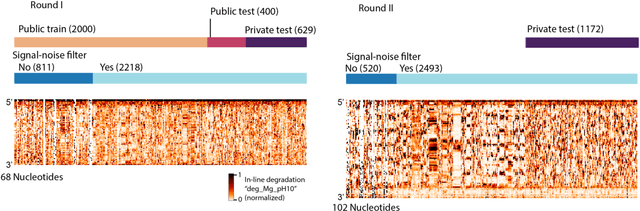
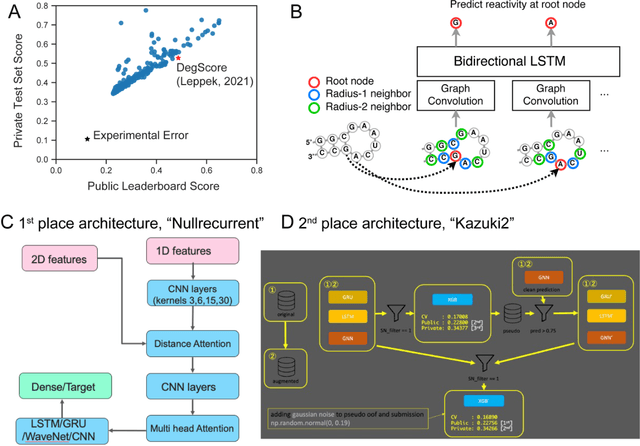
Abstract:Messenger RNA-based medicines hold immense potential, as evidenced by their rapid deployment as COVID-19 vaccines. However, worldwide distribution of mRNA molecules has been limited by their thermostability, which is fundamentally limited by the intrinsic instability of RNA molecules to a chemical degradation reaction called in-line hydrolysis. Predicting the degradation of an RNA molecule is a key task in designing more stable RNA-based therapeutics. Here, we describe a crowdsourced machine learning competition ("Stanford OpenVaccine") on Kaggle, involving single-nucleotide resolution measurements on 6043 102-130-nucleotide diverse RNA constructs that were themselves solicited through crowdsourcing on the RNA design platform Eterna. The entire experiment was completed in less than 6 months. Winning models demonstrated test set errors that were better by 50% than the previous state-of-the-art DegScore model. Furthermore, these models generalized to blindly predicting orthogonal degradation data on much longer mRNA molecules (504-1588 nucleotides) with improved accuracy over DegScore and other models. Top teams integrated natural language processing architectures and data augmentation techniques with predictions from previous dynamic programming models for RNA secondary structure. These results indicate that such models are capable of representing in-line hydrolysis with excellent accuracy, supporting their use for designing stabilized messenger RNAs. The integration of two crowdsourcing platforms, one for data set creation and another for machine learning, may be fruitful for other urgent problems that demand scientific discovery on rapid timescales.
SentRNA: Improving computational RNA design by incorporating a prior of human design strategies
Mar 08, 2018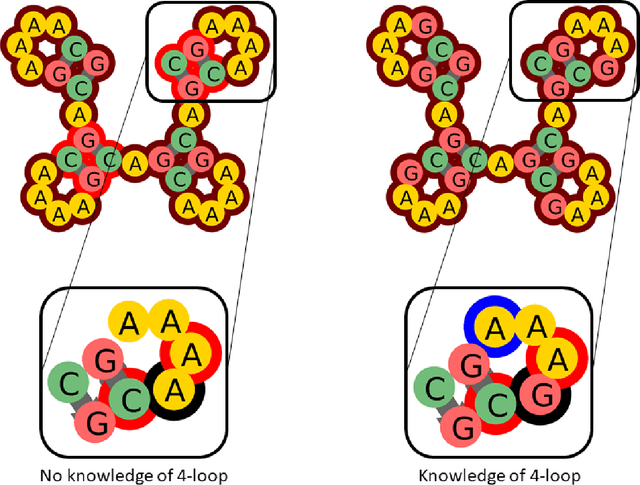
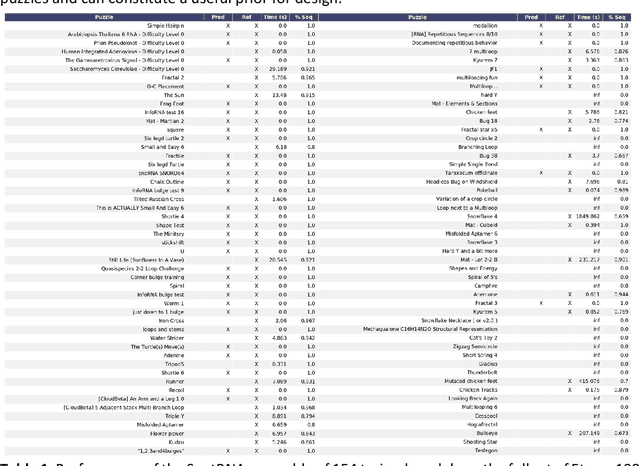
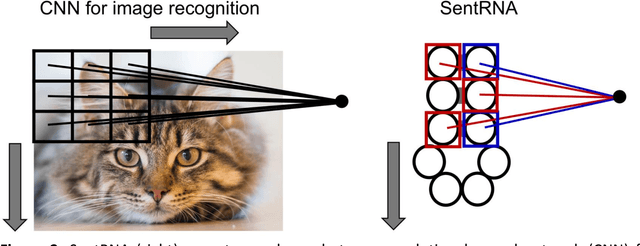
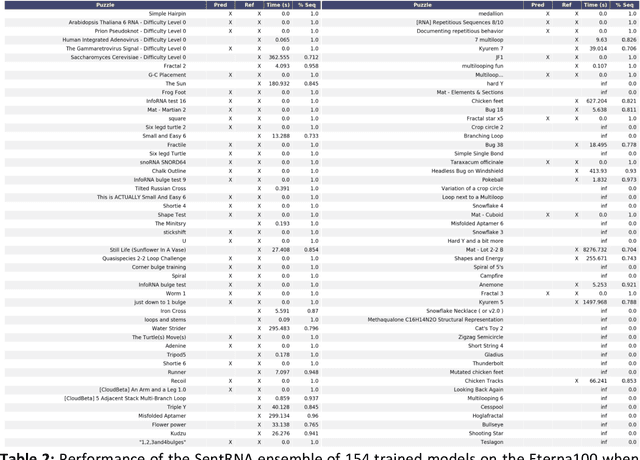
Abstract:Designing RNA sequences that fold into specific structures and perform desired biological functions is an emerging field in bioengineering with broad applications from intracellular chemical catalysis to cancer therapy via selective gene silencing. Effective RNA design requires first solving the inverse folding problem: given a target structure, propose a sequence that folds into that structure. Although significant progress has been made in developing computational algorithms for this purpose, current approaches are ineffective at designing sequences for complex targets, limiting their utility in real-world applications. However, an alternative that has shown significantly higher performance are human players of the online RNA design game EteRNA. Through many rounds of gameplay, these players have developed a collective library of "human" rules and strategies for RNA design that have proven to be more effective than current computational approaches, especially for complex targets. Here, we present an RNA design agent, SentRNA, which consists of a fully-connected neural network trained using the $eternasolves$ dataset, a set of $1.8 x 10^4$ player-submitted sequences across 724 unique targets. The agent first predicts an initial sequence for a target using the trained network, and then refines that solution if necessary using a short adaptive walk utilizing a canon of standard design moves. Through this approach, we observe SentRNA can learn and apply human-like design strategies to solve several complex targets previously unsolvable by any computational approach. We thus demonstrate that incorporating a prior of human design strategies into a computational agent can significantly boost its performance, and suggests a new paradigm for machine-based RNA design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge