Jade Shi
EteRNA players
SentRNA: Improving computational RNA design by incorporating a prior of human design strategies
Mar 08, 2018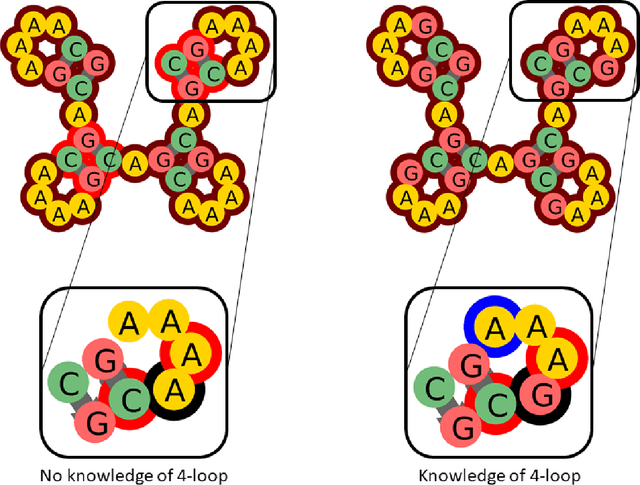
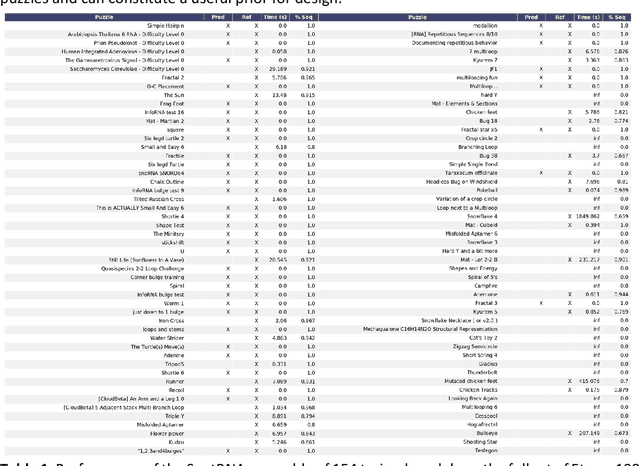
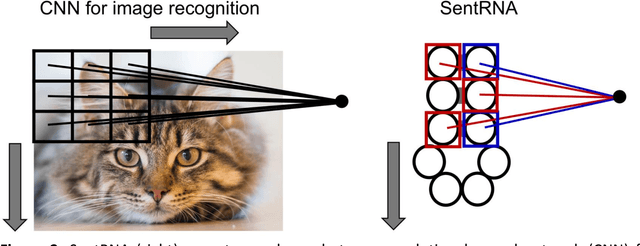
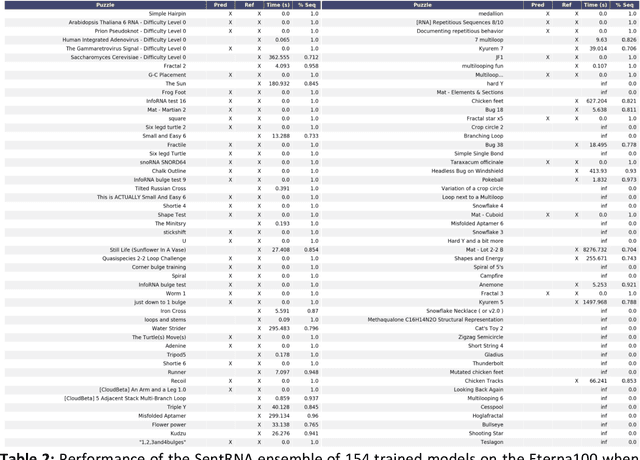
Abstract:Designing RNA sequences that fold into specific structures and perform desired biological functions is an emerging field in bioengineering with broad applications from intracellular chemical catalysis to cancer therapy via selective gene silencing. Effective RNA design requires first solving the inverse folding problem: given a target structure, propose a sequence that folds into that structure. Although significant progress has been made in developing computational algorithms for this purpose, current approaches are ineffective at designing sequences for complex targets, limiting their utility in real-world applications. However, an alternative that has shown significantly higher performance are human players of the online RNA design game EteRNA. Through many rounds of gameplay, these players have developed a collective library of "human" rules and strategies for RNA design that have proven to be more effective than current computational approaches, especially for complex targets. Here, we present an RNA design agent, SentRNA, which consists of a fully-connected neural network trained using the $eternasolves$ dataset, a set of $1.8 x 10^4$ player-submitted sequences across 724 unique targets. The agent first predicts an initial sequence for a target using the trained network, and then refines that solution if necessary using a short adaptive walk utilizing a canon of standard design moves. Through this approach, we observe SentRNA can learn and apply human-like design strategies to solve several complex targets previously unsolvable by any computational approach. We thus demonstrate that incorporating a prior of human design strategies into a computational agent can significantly boost its performance, and suggests a new paradigm for machine-based RNA design.
Retrosynthetic reaction prediction using neural sequence-to-sequence models
Jun 06, 2017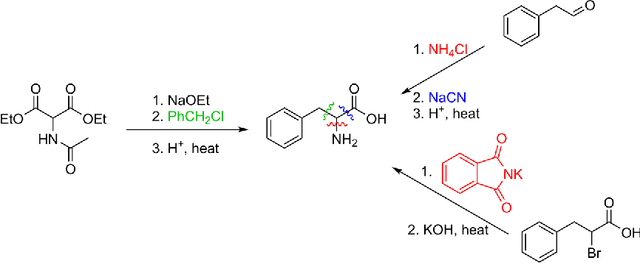
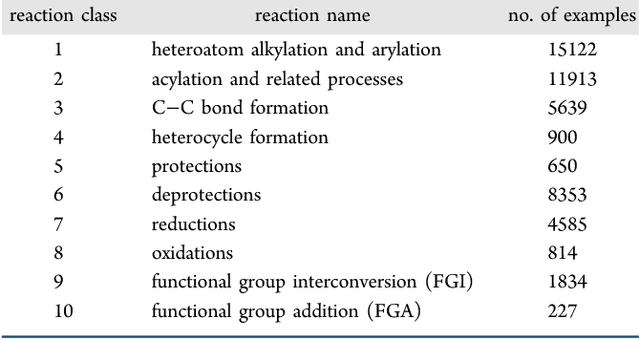
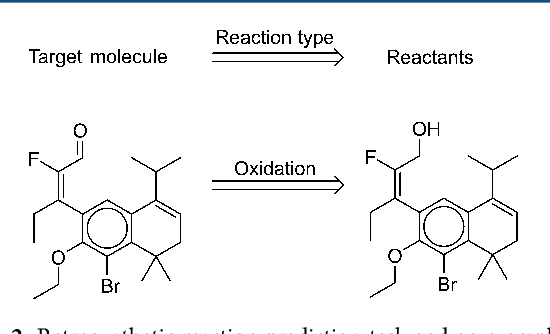
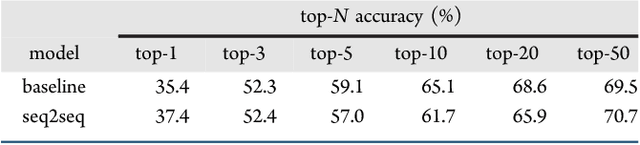
Abstract:We describe a fully data driven model that learns to perform a retrosynthetic reaction prediction task, which is treated as a sequence-to-sequence mapping problem. The end-to-end trained model has an encoder-decoder architecture that consists of two recurrent neural networks, which has previously shown great success in solving other sequence-to-sequence prediction tasks such as machine translation. The model is trained on 50,000 experimental reaction examples from the United States patent literature, which span 10 broad reaction types that are commonly used by medicinal chemists. We find that our model performs comparably with a rule-based expert system baseline model, and also overcomes certain limitations associated with rule-based expert systems and with any machine learning approach that contains a rule-based expert system component. Our model provides an important first step towards solving the challenging problem of computational retrosynthetic analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge