Rahil Mehrizi
Multi-omics Prediction from High-content Cellular Imaging with Deep Learning
Jun 19, 2023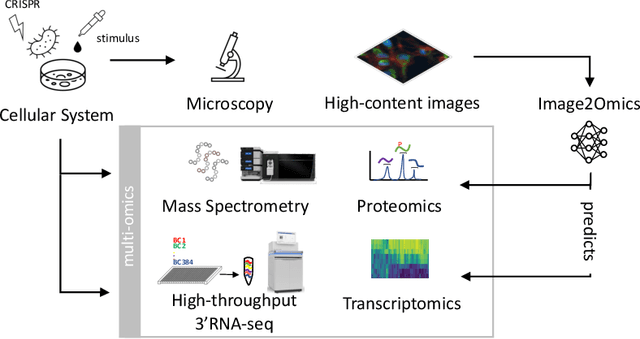
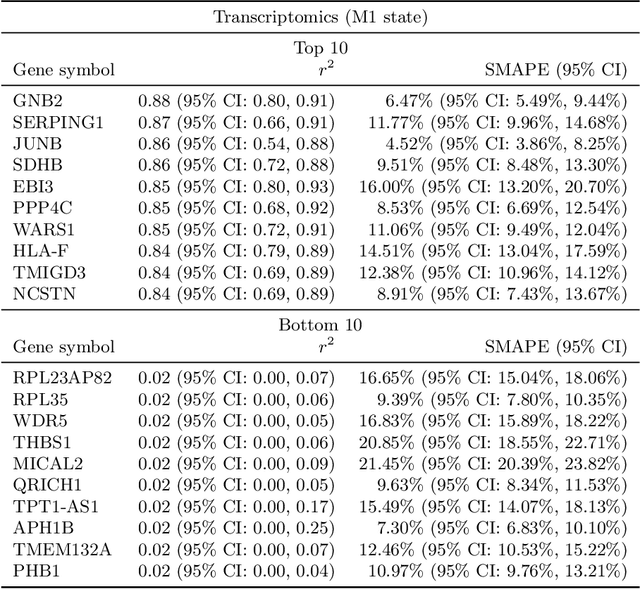
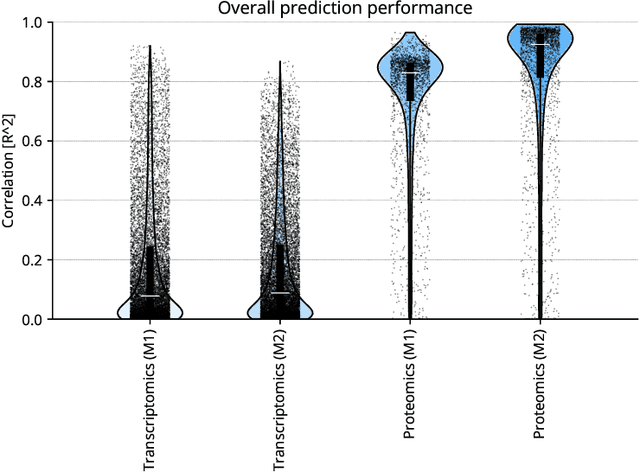
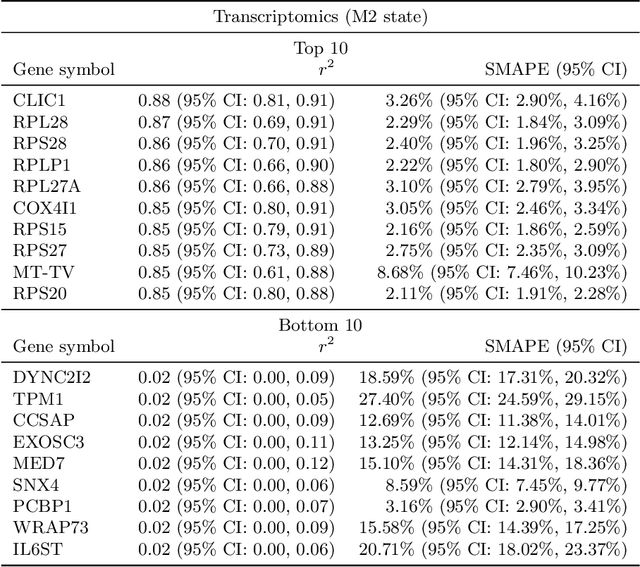
Abstract:High-content cellular imaging, transcriptomics, and proteomics data provide rich and complementary views on the molecular layers of biology that influence cellular states and function. However, the biological determinants through which changes in multi-omics measurements influence cellular morphology have not yet been systematically explored, and the degree to which cell imaging could potentially enable the prediction of multi-omics directly from cell imaging data is therefore currently unclear. Here, we address the question of whether it is possible to predict bulk multi-omics measurements directly from cell images using Image2Omics -- a deep learning approach that predicts multi-omics in a cell population directly from high-content images stained with multiplexed fluorescent dyes. We perform an experimental evaluation in gene-edited macrophages derived from human induced pluripotent stem cell (hiPSC) under multiple stimulation conditions and demonstrate that Image2Omics achieves significantly better performance in predicting transcriptomics and proteomics measurements directly from cell images than predictors based on the mean observed training set abundance. We observed significant predictability of abundances for 5903 (22.43%; 95% CI: 8.77%, 38.88%) and 5819 (22.11%; 95% CI: 10.40%, 38.08%) transcripts out of 26137 in M1 and M2-stimulated macrophages respectively and for 1933 (38.77%; 95% CI: 36.94%, 39.85%) and 2055 (41.22%; 95% CI: 39.31%, 42.42%) proteins out of 4986 in M1 and M2-stimulated macrophages respectively. Our results show that some transcript and protein abundances are predictable from cell imaging and that cell imaging may potentially, in some settings and depending on the mechanisms of interest and desired performance threshold, even be a scalable and resource-efficient substitute for multi-omics measurements.
Automatic Health Problem Detection from Gait Videos Using Deep Neural Networks
Jun 04, 2019



Abstract:The aim of this study is developing an automatic system for detection of gait-related health problems using Deep Neural Networks (DNNs). The proposed system takes a video of patients as the input and estimates their 3D body pose using a DNN based method. Our code is publicly available at https://github.com/rmehrizi/multi-view-pose-estimation. The resulting 3D body pose time series are then analyzed in a classifier, which classifies input gait videos into four different groups including Healthy, with Parkinsons disease, Post Stroke patient, and with orthopedic problems. The proposed system removes the requirement of complex and heavy equipment and large laboratory space, and makes the system practical for home use. Moreover, it does not need domain knowledge for feature engineering since it is capable of extracting semantic and high level features from the input data. The experimental results showed the classification accuracy of 56% to 96% for different groups. Furthermore, only 1 out of 25 healthy subjects were misclassified (False positive), and only 1 out of 70 patients were classified as a healthy subject (False negative). This study presents a starting point toward a powerful tool for automatic classification of gait disorders and can be used as a basis for future applications of Deep Learning in clinical gait analysis. Since the system uses digital cameras as the only required equipment, it can be employed in domestic environment of patients and elderly people for consistent gait monitoring and early detection of gait alterations.
Toward Marker-free 3D Pose Estimation in Lifting: A Deep Multi-view Solution
Feb 06, 2018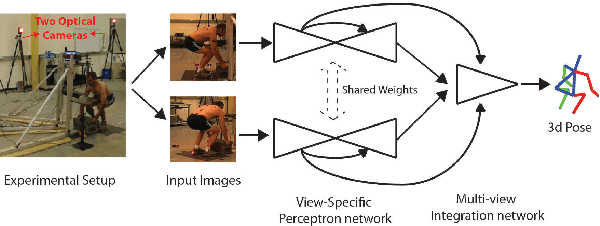
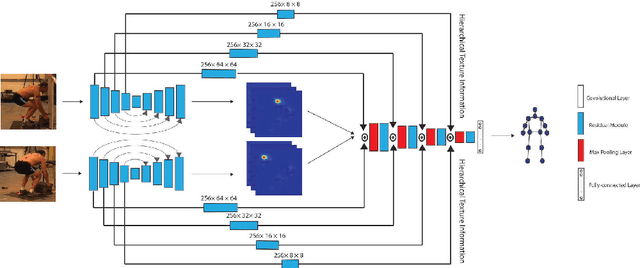
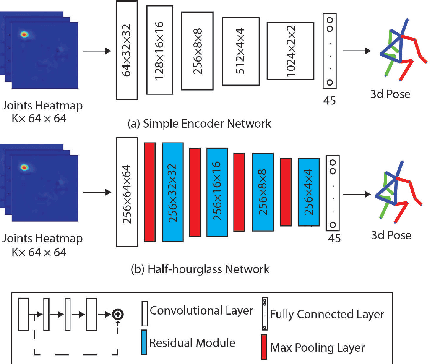
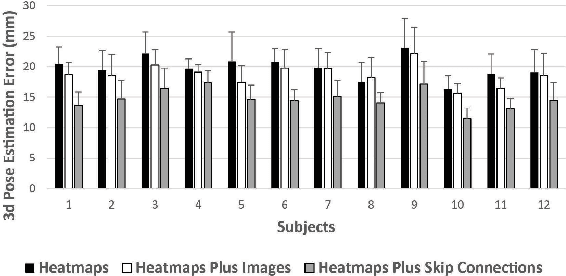
Abstract:Lifting is a common manual material handling task performed in the workplaces. It is considered as one of the main risk factors for Work-related Musculoskeletal Disorders. To improve work place safety, it is necessary to assess musculoskeletal and biomechanical risk exposures associated with these tasks, which requires very accurate 3D pose. Existing approaches mainly utilize marker-based sensors to collect 3D information. However, these methods are usually expensive to setup, time-consuming in process, and sensitive to the surrounding environment. In this study, we propose a multi-view based deep perceptron approach to address aforementioned limitations. Our approach consists of two modules: a "view-specific perceptron" network extracts rich information independently from the image of view, which includes both 2D shape and hierarchical texture information; while a "multi-view integration" network synthesizes information from all available views to predict accurate 3D pose. To fully evaluate our approach, we carried out comprehensive experiments to compare different variants of our design. The results prove that our approach achieves comparable performance with former marker-based methods, i.e. an average error of $14.72 \pm 2.96$ mm on the lifting dataset. The results are also compared with state-of-the-art methods on HumanEva-I dataset, which demonstrates the superior performance of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge