Rafał Kurczab
Pharmacoprint -- a combination of pharmacophore fingerprint and artificial intelligence as a tool for computer-aided drug design
Oct 04, 2021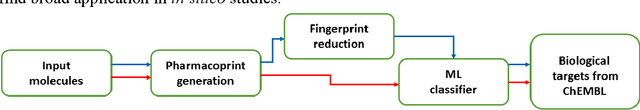
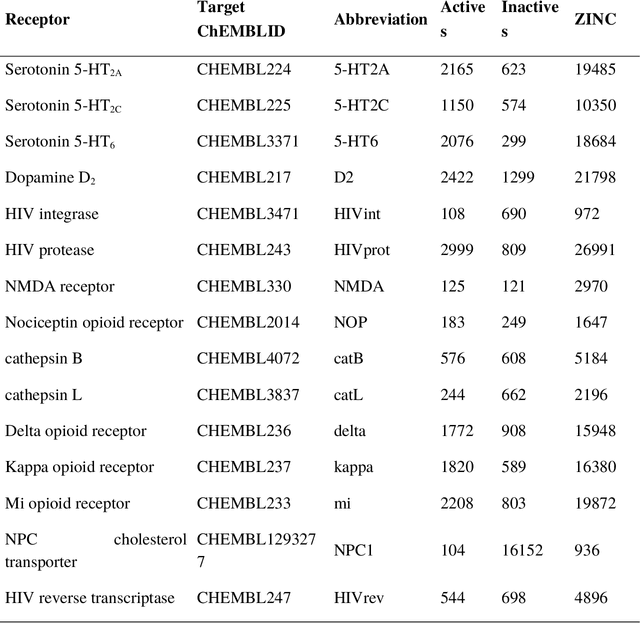
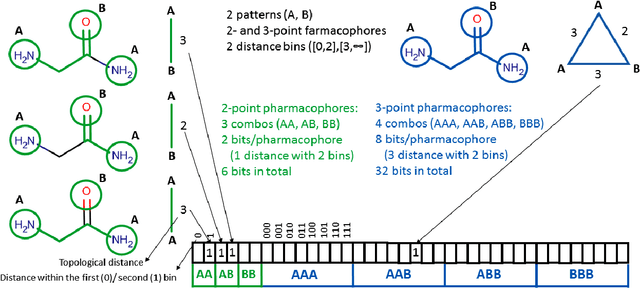
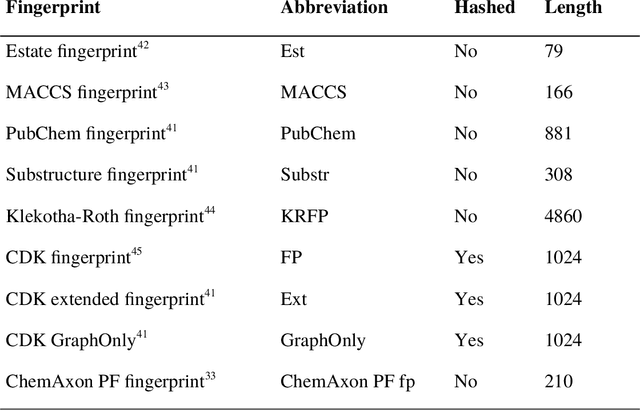
Abstract:Structural fingerprints and pharmacophore modeling are methodologies that have been used for at least two decades in various fields of cheminformatics: from similarity searching to machine learning (ML). Advances in silico techniques consequently led to combining both these methodologies into a new approach known as pharmacophore fingerprint. Herein, we propose a high-resolution, pharmacophore fingerprint called Pharmacoprint that encodes the presence, types, and relationships between pharmacophore features of a molecule. Pharmacoprint was evaluated in classification experiments by using ML algorithms (logistic regression, support vector machines, linear support vector machines, and neural networks) and outperformed other popular molecular fingerprints (i.e., Estate, MACCS, PubChem, Substructure, Klekotha-Roth, CDK, Extended, and GraphOnly) and ChemAxon Pharmacophoric Features fingerprint. Pharmacoprint consisted of 39973 bits; several methods were applied for dimensionality reduction, and the best algorithm not only reduced the length of bit string but also improved the efficiency of ML tests. Further optimization allowed us to define the best parameter settings for using Pharmacoprint in discrimination tests and for maximizing statistical parameters. Finally, Pharmacoprint generated for 3D structures with defined hydrogens as input data was applied to neural networks with a supervised autoencoder for selecting the most important bits and allowed to maximize Matthews Correlation Coefficient up to 0.962. The results show the potential of Pharmacoprint as a new, perspective tool for computer-aided drug design.
PluGeN: Multi-Label Conditional Generation From Pre-Trained Models
Sep 18, 2021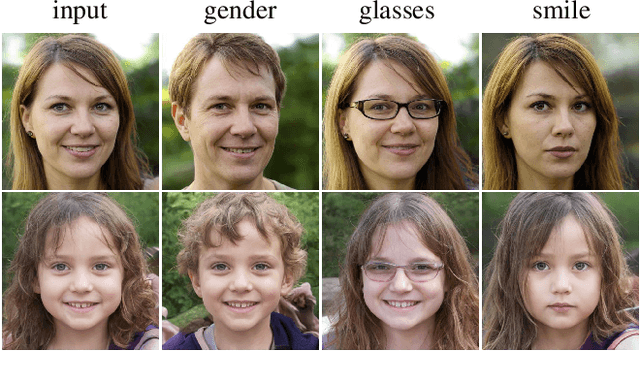
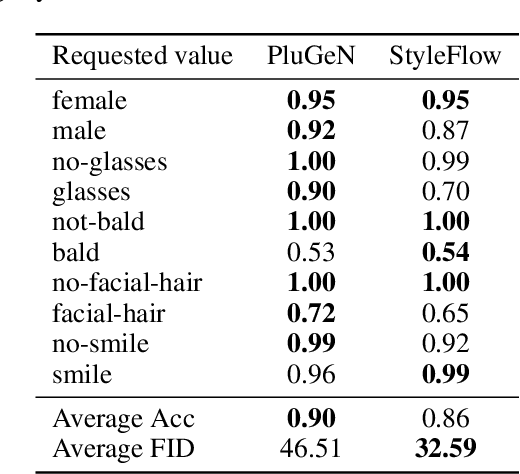
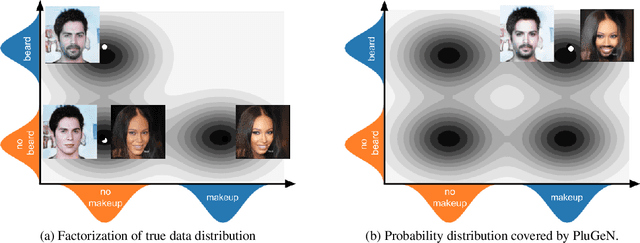
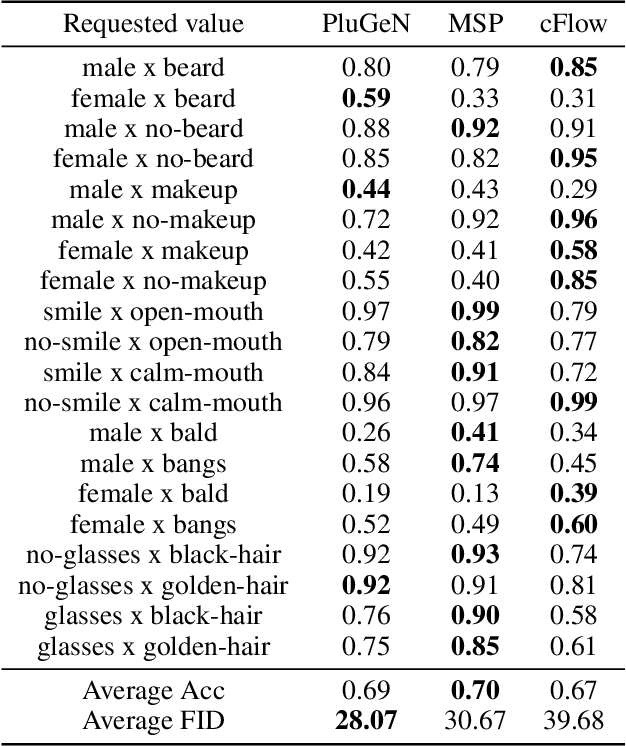
Abstract:Modern generative models achieve excellent quality in a variety of tasks including image or text generation and chemical molecule modeling. However, existing methods often lack the essential ability to generate examples with requested properties, such as the age of the person in the photo or the weight of the generated molecule. Incorporating such additional conditioning factors would require rebuilding the entire architecture and optimizing the parameters from scratch. Moreover, it is difficult to disentangle selected attributes so that to perform edits of only one attribute while leaving the others unchanged. To overcome these limitations we propose PluGeN (Plugin Generative Network), a simple yet effective generative technique that can be used as a plugin to pre-trained generative models. The idea behind our approach is to transform the entangled latent representation using a flow-based module into a multi-dimensional space where the values of each attribute are modeled as an independent one-dimensional distribution. In consequence, PluGeN can generate new samples with desired attributes as well as manipulate labeled attributes of existing examples. Due to the disentangling of the latent representation, we are even able to generate samples with rare or unseen combinations of attributes in the dataset, such as a young person with gray hair, men with make-up, or women with beards. We combined PluGeN with GAN and VAE models and applied it to conditional generation and manipulation of images and chemical molecule modeling. Experiments demonstrate that PluGeN preserves the quality of backbone models while adding the ability to control the values of labeled attributes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge