Prashant Nagpal
Modality-AGnostic Image Cascade (MAGIC) for Multi-Modality Cardiac Substructure Segmentation
Jun 12, 2025Abstract:Cardiac substructures are essential in thoracic radiation therapy planning to minimize risk of radiation-induced heart disease. Deep learning (DL) offers efficient methods to reduce contouring burden but lacks generalizability across different modalities and overlapping structures. This work introduces and validates a Modality-AGnostic Image Cascade (MAGIC) for comprehensive and multi-modal cardiac substructure segmentation. MAGIC is implemented through replicated encoding and decoding branches of an nnU-Net-based, U-shaped backbone conserving the function of a single model. Twenty cardiac substructures (heart, chambers, great vessels (GVs), valves, coronary arteries (CAs), and conduction nodes) from simulation CT (Sim-CT), low-field MR-Linac, and cardiac CT angiography (CCTA) modalities were manually delineated and used to train (n=76), validate (n=15), and test (n=30) MAGIC. Twelve comparison models (four segmentation subgroups across three modalities) were equivalently trained. All methods were compared for training efficiency and against reference contours using the Dice Similarity Coefficient (DSC) and two-tailed Wilcoxon Signed-Rank test (threshold, p<0.05). Average DSC scores were 0.75(0.16) for Sim-CT, 0.68(0.21) for MR-Linac, and 0.80(0.16) for CCTA. MAGIC outperforms the comparison in 57% of cases, with limited statistical differences. MAGIC offers an effective and accurate segmentation solution that is lightweight and capable of segmenting multiple modalities and overlapping structures in a single model. MAGIC further enables clinical implementation by simplifying the computational requirements and offering unparalleled flexibility for clinical settings.
Joint alignment and reconstruction of multislice dynamic MRI using variational manifold learning
Nov 21, 2021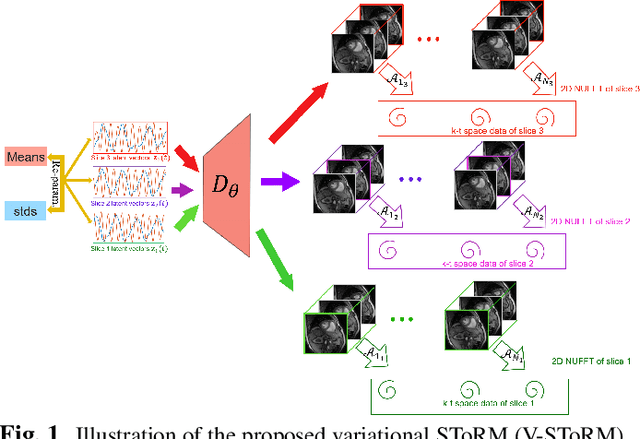
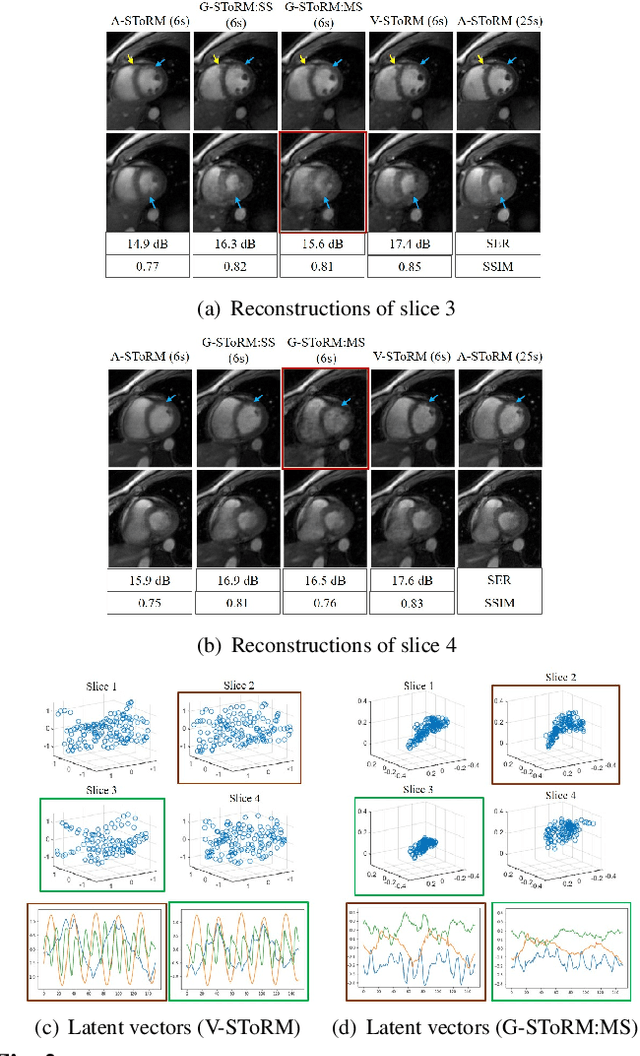

Abstract:Free-breathing cardiac MRI schemes are emerging as competitive alternatives to breath-held cine MRI protocols, enabling applicability to pediatric and other population groups that cannot hold their breath. Because the data from the slices are acquired sequentially, the cardiac/respiratory motion patterns may be different for each slice; current free-breathing approaches perform independent recovery of each slice. In addition to not being able to exploit the inter-slice redundancies, manual intervention or sophisticated post-processing methods are needed to align the images post-recovery for quantification. To overcome these challenges, we propose an unsupervised variational deep manifold learning scheme for the joint alignment and reconstruction of multislice dynamic MRI. The proposed scheme jointly learns the parameters of the deep network as well as the latent vectors for each slice, which capture the motion-induced dynamic variations, from the k-t space data of the specific subject. The variational framework minimizes the non-uniqueness in the representation, thus offering improved alignment and reconstructions.
Dynamic Imaging using Deep Bi-linear Unsupervised Regularization (DEBLUR)
Jun 30, 2021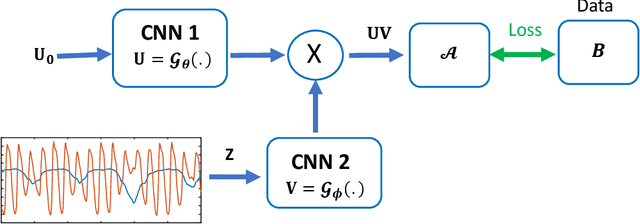
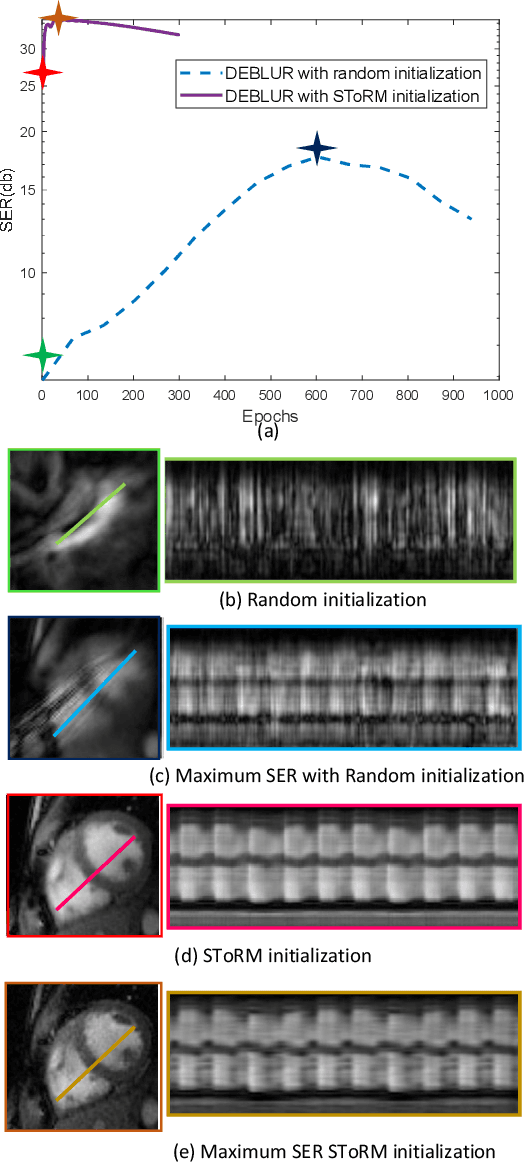

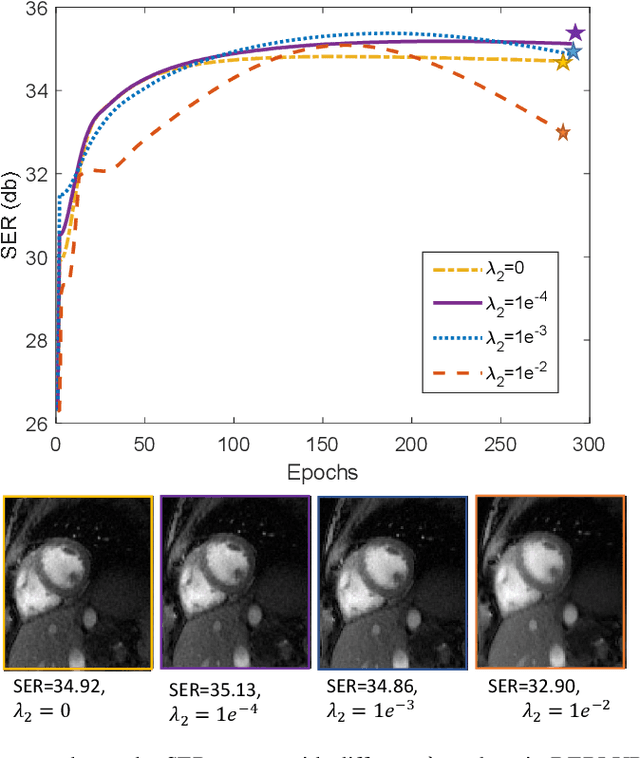
Abstract:Bilinear models that decompose dynamic data to spatial and temporal factors are powerful and memory-efficient tools for the recovery of dynamic MRI data. These methods rely on sparsity and energy compaction priors on the factors to regularize the recovery. The quality of the recovered images depend on the specific priors. Motivated by deep image prior, we introduce a novel bilinear model whose factors are represented using convolutional neural networks (CNNs). The CNN parameters are learned from the undersampled data off the same subject. To reduce the run time and to improve performance, we initialize the CNN parameters. We use sparsity regularization of the network parameters to minimize the overfitting of the network to measurement noise. Our experiments on free breathing and ungated cardiac cine data acquired using a navigated golden-angle gradient-echo radial sequence show the ability of our method to provide reduced spatial blurring as compared to low-rank and SToRM reconstructions.
Dynamic imaging using a deep generative SToRM model
Feb 11, 2021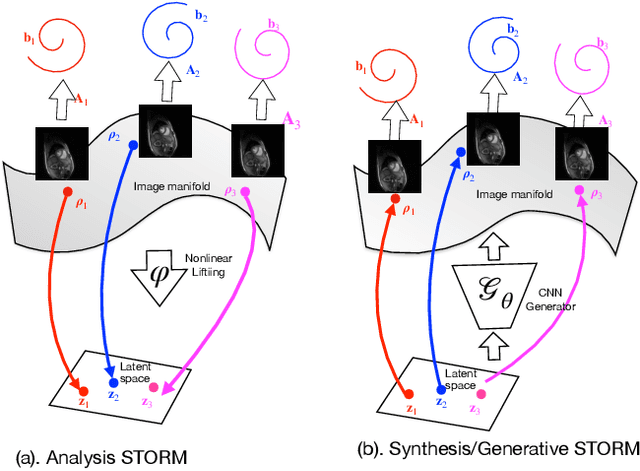
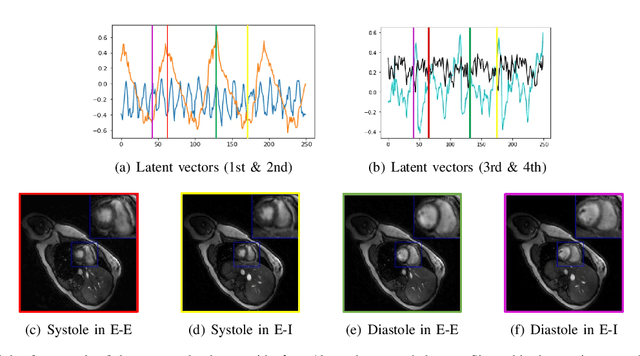
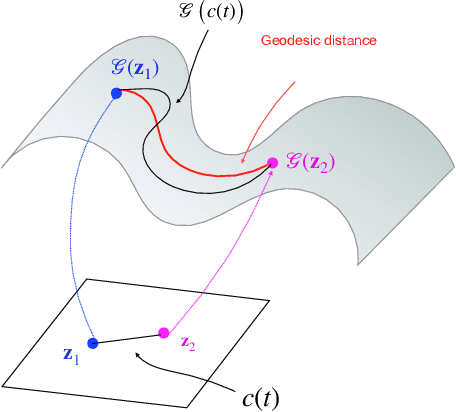
Abstract:We introduce a generative smoothness regularization on manifolds (SToRM) model for the recovery of dynamic image data from highly undersampled measurements. The model assumes that the images in the dataset are non-linear mappings of low-dimensional latent vectors. We use the deep convolutional neural network (CNN) to represent the non-linear transformation. The parameters of the generator as well as the low-dimensional latent vectors are jointly estimated only from the undersampled measurements. This approach is different from traditional CNN approaches that require extensive fully sampled training data. We penalize the norm of the gradients of the non-linear mapping to constrain the manifold to be smooth, while temporal gradients of the latent vectors are penalized to obtain a smoothly varying time-series. The proposed scheme brings in the spatial regularization provided by the convolutional network. The main benefit of the proposed scheme is the improvement in image quality and the orders-of-magnitude reduction in memory demand compared to traditional manifold models. To minimize the computational complexity of the algorithm, we introduce an efficient progressive training-in-time approach and an approximate cost function. These approaches speed up the image reconstructions and offers better reconstruction performance.
Deep Generative SToRM model for dynamic imaging
Jan 29, 2021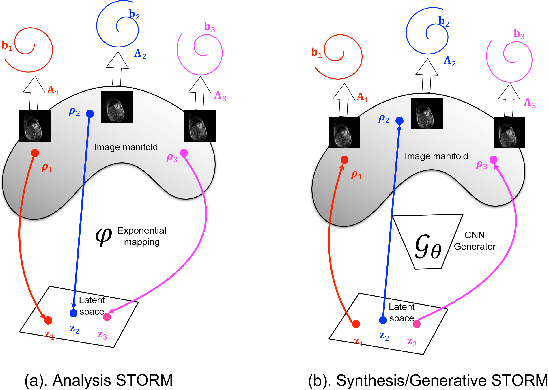
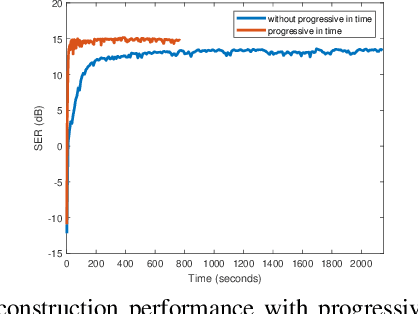
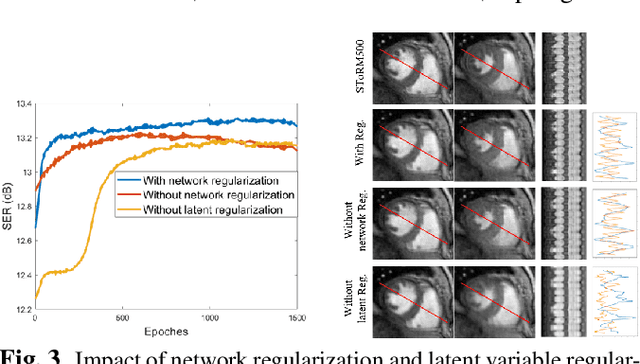
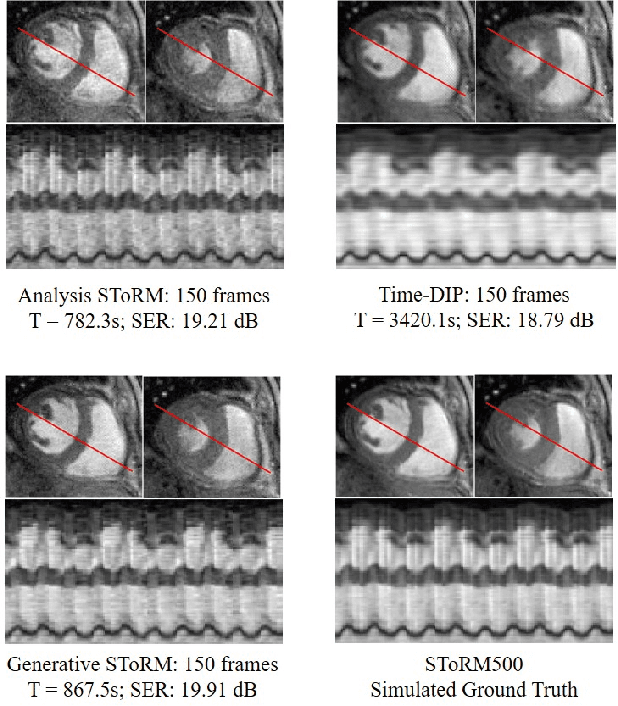
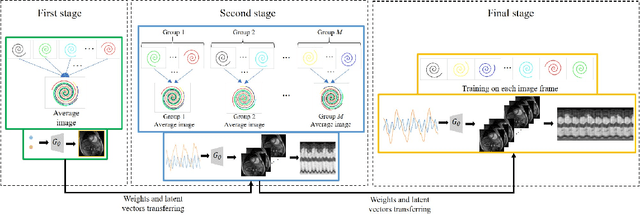
Abstract:We introduce a novel generative smoothness regularization on manifolds (SToRM) model for the recovery of dynamic image data from highly undersampled measurements. The proposed generative framework represents the image time series as a smooth non-linear function of low-dimensional latent vectors that capture the cardiac and respiratory phases. The non-linear function is represented using a deep convolutional neural network (CNN). Unlike the popular CNN approaches that require extensive fully-sampled training data that is not available in this setting, the parameters of the CNN generator as well as the latent vectors are jointly estimated from the undersampled measurements using stochastic gradient descent. We penalize the norm of the gradient of the generator to encourage the learning of a smooth surface/manifold, while temporal gradients of the latent vectors are penalized to encourage the time series to be smooth. The main benefits of the proposed scheme are (a) the quite significant reduction in memory demand compared to the analysis based SToRM model, and (b) the spatial regularization brought in by the CNN model. We also introduce efficient progressive approaches to minimize the computational complexity of the algorithm.
Alignment & joint recovery of multi-slice dynamic MRI using deep generative manifold model
Jan 20, 2021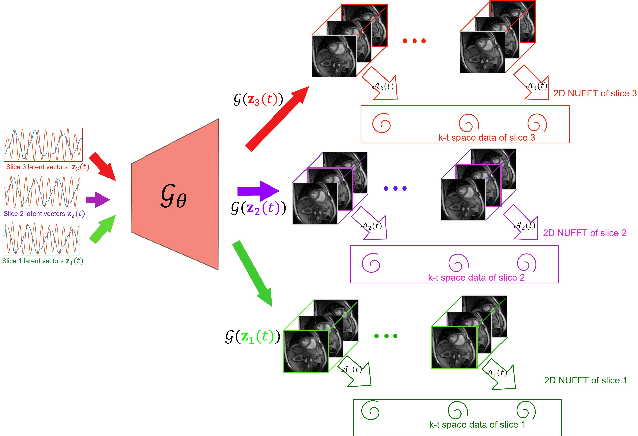
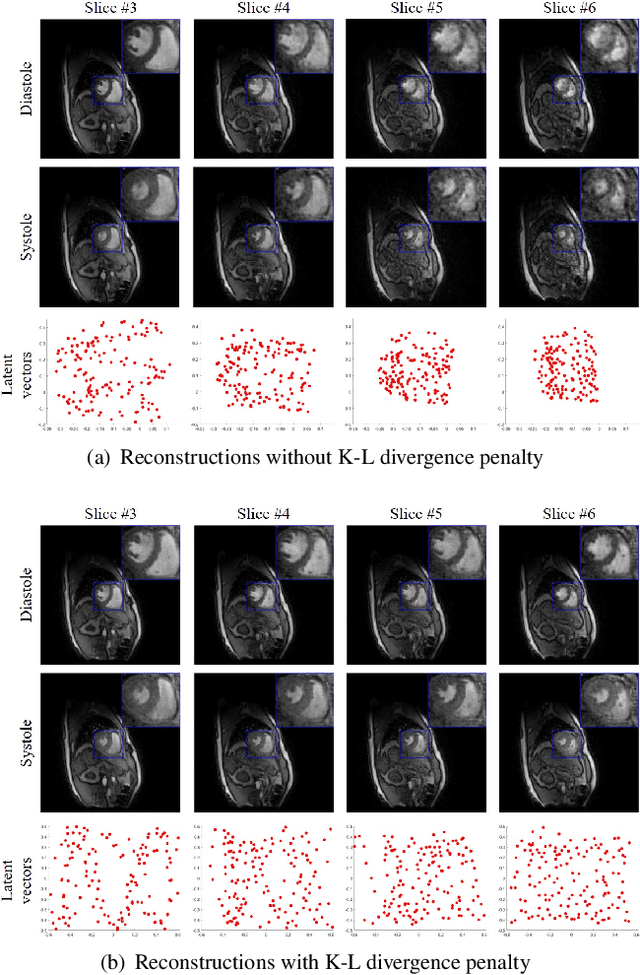
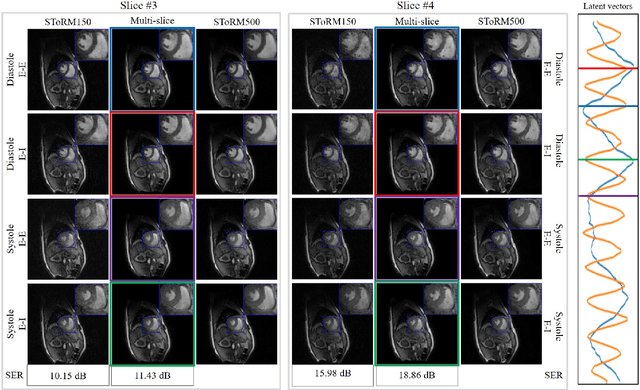
Abstract:We introduce a novel unsupervised deep generative manifold model for the recovery of multi-slice free-breathing and ungated cardiac MRI from highly undersampled measurements. The proposed scheme represents the multi-slice volume at each time point as the output of a deep convolutional neural network (CNN) generator, which is driven by latent vectors that capture the cardiac and respiratory phase at the specific time point. The main difference between the proposed method and the traditional CNN approaches is that the proposed scheme learns the network parameters from only the highly undersampled data rather than the extensive fully-sampled training data. We also learn the latent codes from the undersampled data using the stochastic gradient descent. Regularizations on the network and the latent codes are introduced to encourage the learning of smooth image manifold and the latent codes for each slice have the same distribution. The main benefits of the proposed scheme are (a) the ability to align multi-slice data and capitalize on the redundancy between the slices; (b) the ability to estimate the gating information directly from the k-t space data; and (c) the unsupervised learning strategy that eliminates the need for extensive training data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge