Po-Chen Kuo
Tractable Dendritic RNNs for Reconstructing Nonlinear Dynamical Systems
Jul 06, 2022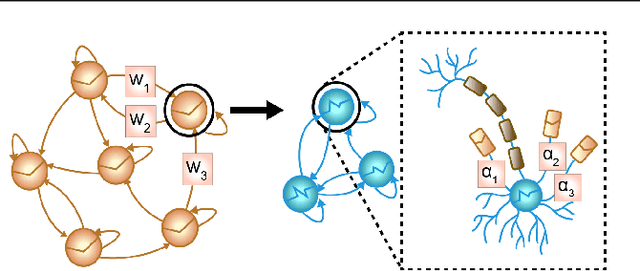
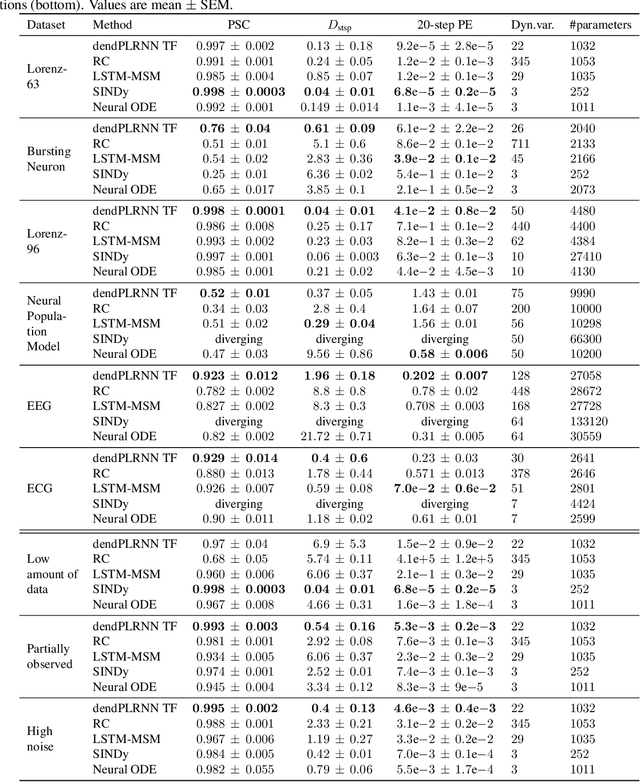
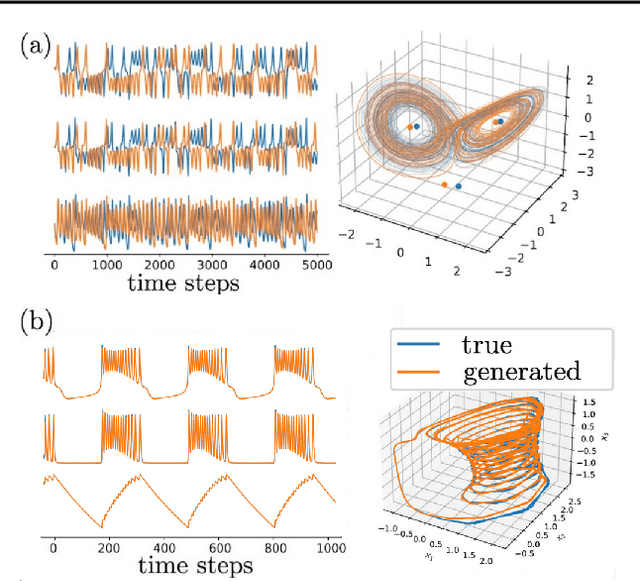
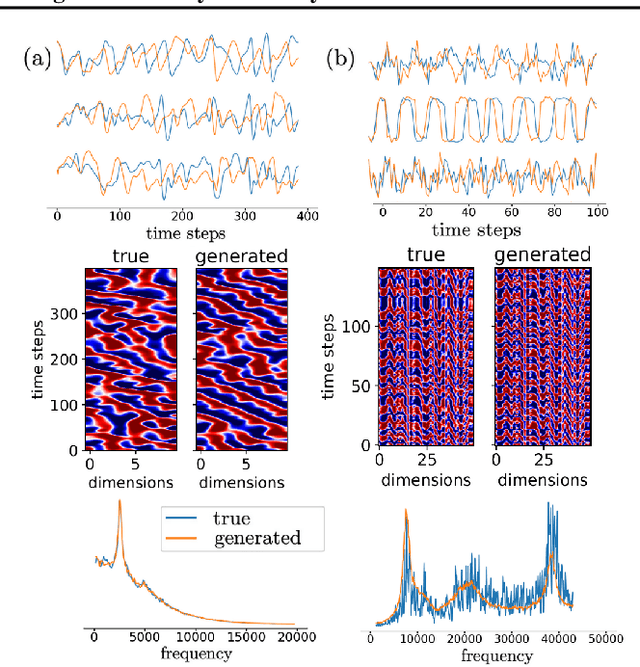
Abstract:In many scientific disciplines, we are interested in inferring the nonlinear dynamical system underlying a set of observed time series, a challenging task in the face of chaotic behavior and noise. Previous deep learning approaches toward this goal often suffered from a lack of interpretability and tractability. In particular, the high-dimensional latent spaces often required for a faithful embedding, even when the underlying dynamics lives on a lower-dimensional manifold, can hamper theoretical analysis. Motivated by the emerging principles of dendritic computation, we augment a dynamically interpretable and mathematically tractable piecewise-linear (PL) recurrent neural network (RNN) by a linear spline basis expansion. We show that this approach retains all the theoretically appealing properties of the simple PLRNN, yet boosts its capacity for approximating arbitrary nonlinear dynamical systems in comparatively low dimensions. We employ two frameworks for training the system, one combining back-propagation-through-time (BPTT) with teacher forcing, and another based on fast and scalable variational inference. We show that the dendritically expanded PLRNN achieves better reconstructions with fewer parameters and dimensions on various dynamical systems benchmarks and compares favorably to other methods, while retaining a tractable and interpretable structure.
Self-similarity Student for Partial Label Histopathology Image Segmentation
Jul 19, 2020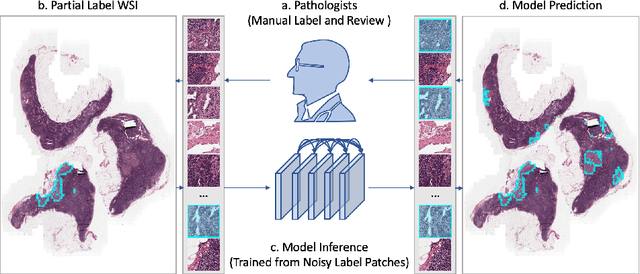
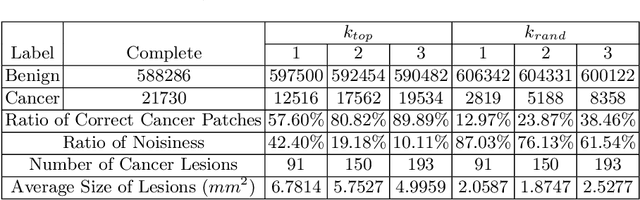
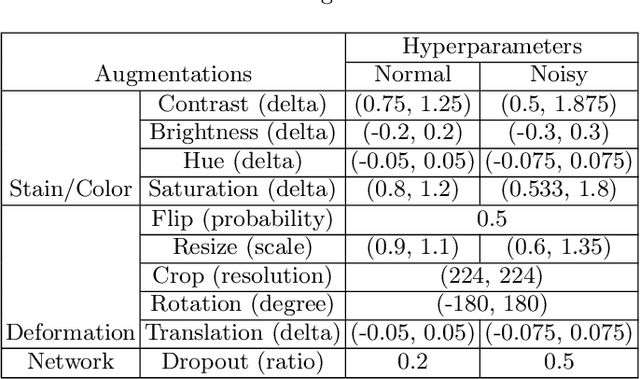
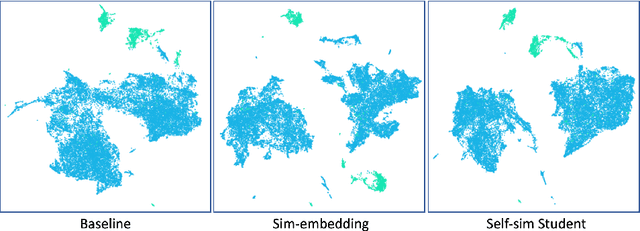
Abstract:Delineation of cancerous regions in gigapixel whole slide images (WSIs) is a crucial diagnostic procedure in digital pathology. This process is time-consuming because of the large search space in the gigapixel WSIs, causing chances of omission and misinterpretation at indistinct tumor lesions. To tackle this, the development of an automated cancerous region segmentation method is imperative. We frame this issue as a modeling problem with partial label WSIs, where some cancerous regions may be misclassified as benign and vice versa, producing patches with noisy labels. To learn from these patches, we propose Self-similarity Student, combining teacher-student model paradigm with similarity learning. Specifically, for each patch, we first sample its similar and dissimilar patches according to spatial distance. A teacher-student model is then introduced, featuring the exponential moving average on both student model weights and teacher predictions ensemble. While our student model takes patches, teacher model takes all their corresponding similar and dissimilar patches for learning robust representation against noisy label patches. Following this similarity learning, our similarity ensemble merges similar patches' ensembled predictions as the pseudo-label of a given patch to counteract its noisy label. On the CAMELYON16 dataset, our method substantially outperforms state-of-the-art noise-aware learning methods by 5$\%$ and the supervised-trained baseline by 10$\%$ in various degrees of noise. Moreover, our method is superior to the baseline on our TVGH TURP dataset with 2$\%$ improvement, demonstrating the generalizability to more clinical histopathology segmentation tasks.
A Cascaded Learning Strategy for Robust COVID-19 Pneumonia Chest X-Ray Screening
Apr 30, 2020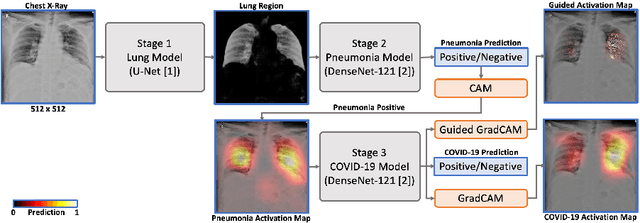
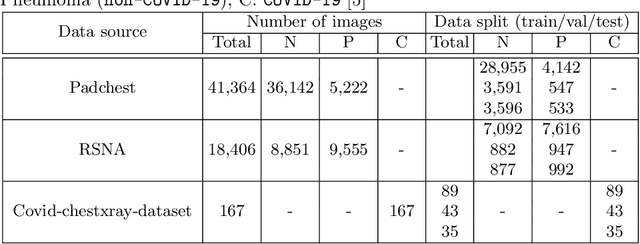
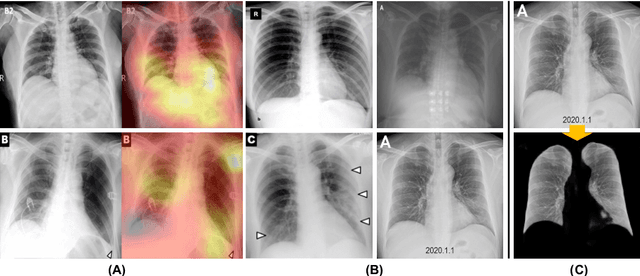
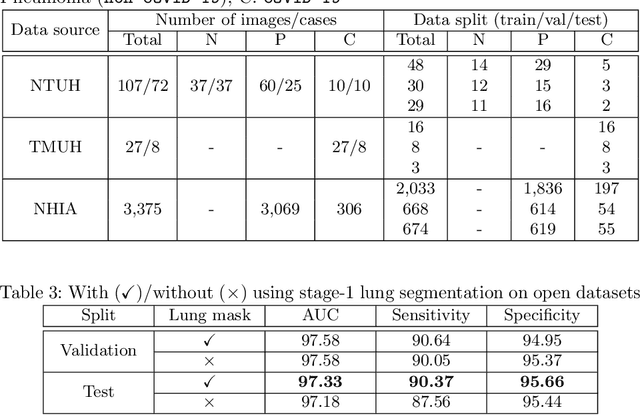
Abstract:We introduce a comprehensive screening platform for the COVID-19 (a.k.a., SARS-CoV-2) pneumonia. The proposed AI-based system works on chest x-ray (CXR) images to predict whether a patient is infected with the COVID-19 disease. Although the recent international joint effort on making the availability of all sorts of open data, the public collection of CXR images is still relatively small for reliably training a deep neural network (DNN) to carry out COVID-19 prediction. To better address such inefficiency, we design a cascaded learning strategy to improve both the sensitivity and the specificity of the resulting DNN classification model. Our approach leverages a large CXR image dataset of non-COVID-19 pneumonia to generalize the original well-trained classification model via a cascaded learning scheme. The resulting screening system is shown to achieve good classification performance on the expanded dataset, including those newly added COVID-19 CXR images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge