Peter Watkinson
DynaGraph: Interpretable Multi-Label Prediction from EHRs via Dynamic Graph Learning and Contrastive Augmentation
Mar 28, 2025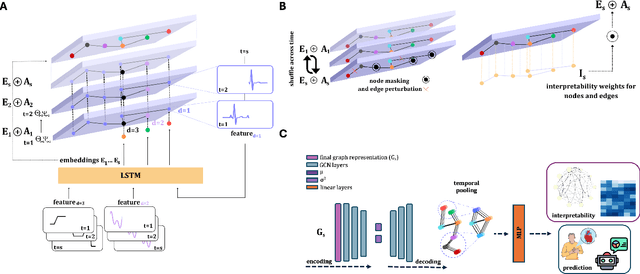
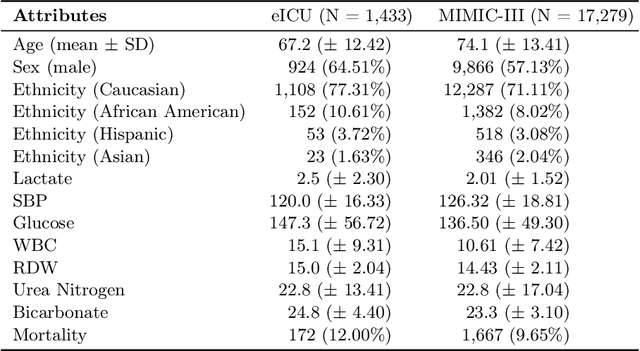
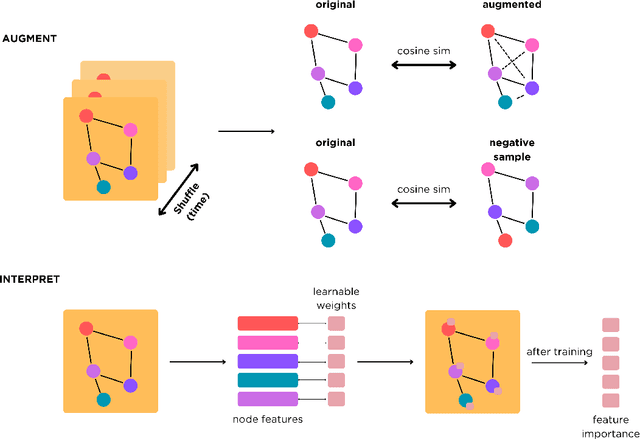
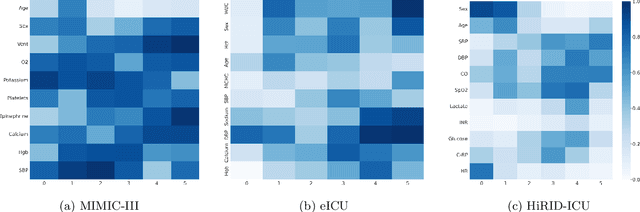
Abstract:Learning from longitudinal electronic health records is limited if it does not capture the temporal trajectories of the patient's state in a clinical setting. Graph models allow us to capture the hidden dependencies of the multivariate time-series when the graphs are constructed in a similar dynamic manner. Previous dynamic graph models require a pre-defined and/or static graph structure, which is unknown in most cases, or they only capture the spatial relations between the features. Furthermore in healthcare, the interpretability of the model is an essential requirement to build trust with clinicians. In addition to previously proposed attention mechanisms, there has not been an interpretable dynamic graph framework for data from multivariate electronic health records (EHRs). Here, we propose DynaGraph, an end-to-end interpretable contrastive graph model that learns the dynamics of multivariate time-series EHRs as part of optimisation. We validate our model in four real-world clinical datasets, ranging from primary care to secondary care settings with broad demographics, in challenging settings where tasks are imbalanced and multi-labelled. Compared to state-of-the-art models, DynaGraph achieves significant improvements in balanced accuracy and sensitivity over the nearest complex competitors in time-series or dynamic graph modelling across three ICU and one primary care datasets. Through a pseudo-attention approach to graph construction, our model also indicates the importance of clinical covariates over time, providing means for clinical validation.
Reinforcement Learning in Dynamic Treatment Regimes Needs Critical Reexamination
May 28, 2024



Abstract:In the rapidly changing healthcare landscape, the implementation of offline reinforcement learning (RL) in dynamic treatment regimes (DTRs) presents a mix of unprecedented opportunities and challenges. This position paper offers a critical examination of the current status of offline RL in the context of DTRs. We argue for a reassessment of applying RL in DTRs, citing concerns such as inconsistent and potentially inconclusive evaluation metrics, the absence of naive and supervised learning baselines, and the diverse choice of RL formulation in existing research. Through a case study with more than 17,000 evaluation experiments using a publicly available Sepsis dataset, we demonstrate that the performance of RL algorithms can significantly vary with changes in evaluation metrics and Markov Decision Process (MDP) formulations. Surprisingly, it is observed that in some instances, RL algorithms can be surpassed by random baselines subjected to policy evaluation methods and reward design. This calls for more careful policy evaluation and algorithm development in future DTR works. Additionally, we discussed potential enhancements toward more reliable development of RL-based dynamic treatment regimes and invited further discussion within the community. Code is available at https://github.com/GilesLuo/ReassessDTR.
DySurv: Dynamic Deep Learning Model for Survival Prediction in the ICU
Oct 28, 2023



Abstract:Survival analysis helps approximate underlying distributions of time-to-events which in the case of critical care like in the ICU can be a powerful tool for dynamic mortality risk prediction. Extending beyond the classical Cox model, deep learning techniques have been leveraged over the last years relaxing the many constraints of their counterparts from statistical methods. In this work, we propose a novel conditional variational autoencoder-based method called DySurv which uses a combination of static and time-series measurements from patient electronic health records in estimating risk of death dynamically in the ICU. DySurv has been tested on standard benchmarks where it outperforms most existing methods including other deep learning methods and we evaluate it on a real-world patient database from MIMIC-IV. The predictive capacity of DySurv is consistent and the survival estimates remain disentangled across different datasets supporting the idea that dynamic deep learning models based on conditional variational inference in multi-task cases can be robust models for survival analysis.
Explainable AI for clinical risk prediction: a survey of concepts, methods, and modalities
Aug 16, 2023Abstract:Recent advancements in AI applications to healthcare have shown incredible promise in surpassing human performance in diagnosis and disease prognosis. With the increasing complexity of AI models, however, concerns regarding their opacity, potential biases, and the need for interpretability. To ensure trust and reliability in AI systems, especially in clinical risk prediction models, explainability becomes crucial. Explainability is usually referred to as an AI system's ability to provide a robust interpretation of its decision-making logic or the decisions themselves to human stakeholders. In clinical risk prediction, other aspects of explainability like fairness, bias, trust, and transparency also represent important concepts beyond just interpretability. In this review, we address the relationship between these concepts as they are often used together or interchangeably. This review also discusses recent progress in developing explainable models for clinical risk prediction, highlighting the importance of quantitative and clinical evaluation and validation across multiple common modalities in clinical practice. It emphasizes the need for external validation and the combination of diverse interpretability methods to enhance trust and fairness. Adopting rigorous testing, such as using synthetic datasets with known generative factors, can further improve the reliability of explainability methods. Open access and code-sharing resources are essential for transparency and reproducibility, enabling the growth and trustworthiness of explainable research. While challenges exist, an end-to-end approach to explainability in clinical risk prediction, incorporating stakeholders from clinicians to developers, is essential for success.
XMI-ICU: Explainable Machine Learning Model for Pseudo-Dynamic Prediction of Mortality in the ICU for Heart Attack Patients
May 10, 2023Abstract:Heart attack remain one of the greatest contributors to mortality in the United States and globally. Patients admitted to the intensive care unit (ICU) with diagnosed heart attack (myocardial infarction or MI) are at higher risk of death. In this study, we use two retrospective cohorts extracted from the eICU and MIMIC-IV databases, to develop a novel pseudo-dynamic machine learning framework for mortality prediction in the ICU with interpretability and clinical risk analysis. The method provides accurate prediction for ICU patients up to 24 hours before the event and provide time-resolved interpretability results. The performance of the framework relying on extreme gradient boosting was evaluated on a held-out test set from eICU, and externally validated on the MIMIC-IV cohort using the most important features identified by time-resolved Shapley values achieving AUCs of 91.0 (balanced accuracy of 82.3) for 6-hour prediction of mortality respectively. We show that our framework successfully leverages time-series physiological measurements by translating them into stacked static prediction problems to be robustly predictive through time in the ICU stay and can offer clinical insight from time-resolved interpretability
Phenotyping Clusters of Patient Trajectories suffering from Chronic Complex Disease
Nov 17, 2020



Abstract:Recent years have seen an increased focus into the tasks of predicting hospital inpatient risk of deterioration and trajectory evolution due to the availability of electronic patient data. A common approach to these problems involves clustering patients time-series information such as vital sign observations) to determine dissimilar subgroups of the patient population. Most clustering methods assume time-invariance of vital-signs and are unable to provide interpretability in clusters that is clinically relevant, for instance, event or outcome information. In this work, we evaluate three different clustering models on a large hospital dataset of vital-sign observations from patients suffering from Chronic Obstructive Pulmonary Disease. We further propose novel modifications to deal with unevenly sampled time-series data and unbalanced class distribution to improve phenotype separation. Lastly, we discuss further avenues of investigation for models to learn patient subgroups with distinct behaviour and phenotype.
Student-Teacher Curriculum Learning via Reinforcement Learning: Predicting Hospital Inpatient Admission Location
Jul 01, 2020



Abstract:Accurate and reliable prediction of hospital admission location is important due to resource-constraints and space availability in a clinical setting, particularly when dealing with patients who come from the emergency department. In this work we propose a student-teacher network via reinforcement learning to deal with this specific problem. A representation of the weights of the student network is treated as the state and is fed as an input to the teacher network. The teacher network's action is to select the most appropriate batch of data to train the student network on from a training set sorted according to entropy. By validating on three datasets, not only do we show that our approach outperforms state-of-the-art methods on tabular data and performs competitively on image recognition, but also that novel curricula are learned by the teacher network. We demonstrate experimentally that the teacher network can actively learn about the student network and guide it to achieve better performance than if trained alone.
* 16 pages, 31 figures, In Proceedings of the 37th International Conference on Machine Learning
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge