Peter V. Coveney
Fast-Forward Lattice Boltzmann: Learning Kinetic Behaviour with Physics-Informed Neural Operators
Sep 26, 2025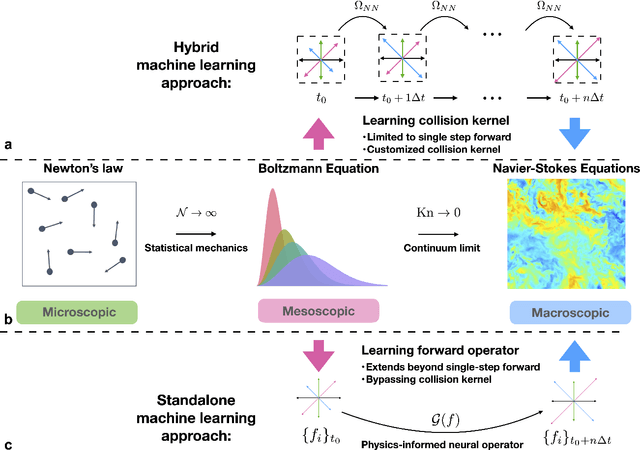
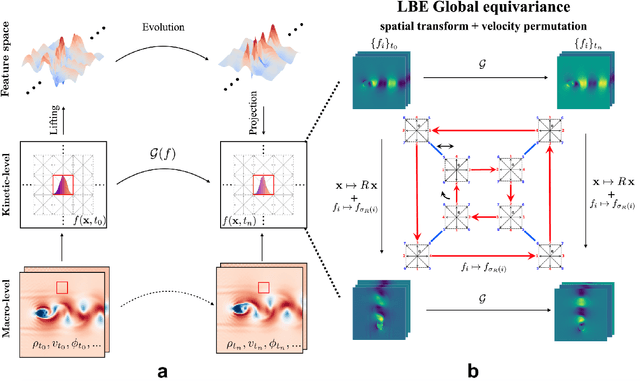
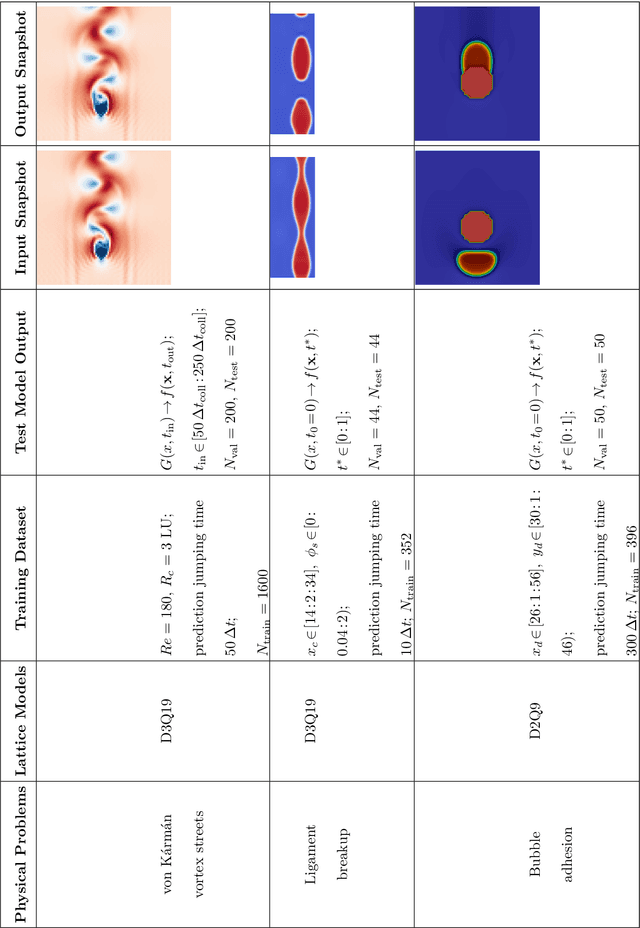
Abstract:The lattice Boltzmann equation (LBE), rooted in kinetic theory, provides a powerful framework for capturing complex flow behaviour by describing the evolution of single-particle distribution functions (PDFs). Despite its success, solving the LBE numerically remains computationally intensive due to strict time-step restrictions imposed by collision kernels. Here, we introduce a physics-informed neural operator framework for the LBE that enables prediction over large time horizons without step-by-step integration, effectively bypassing the need to explicitly solve the collision kernel. We incorporate intrinsic moment-matching constraints of the LBE, along with global equivariance of the full distribution field, enabling the model to capture the complex dynamics of the underlying kinetic system. Our framework is discretization-invariant, enabling models trained on coarse lattices to generalise to finer ones (kinetic super-resolution). In addition, it is agnostic to the specific form of the underlying collision model, which makes it naturally applicable across different kinetic datasets regardless of the governing dynamics. Our results demonstrate robustness across complex flow scenarios, including von Karman vortex shedding, ligament breakup, and bubble adhesion. This establishes a new data-driven pathway for modelling kinetic systems.
A Parameter-Efficient Quantum Anomaly Detection Method on a Superconducting Quantum Processor
Dec 22, 2024



Abstract:Quantum machine learning has gained attention for its potential to address computational challenges. However, whether those algorithms can effectively solve practical problems and outperform their classical counterparts, especially on current quantum hardware, remains a critical question. In this work, we propose a novel quantum machine learning method, called Quantum Support Vector Data Description (QSVDD), for practical anomaly detection, which aims to achieve both parameter efficiency and superior accuracy compared to classical models. Emulation results indicate that QSVDD demonstrates favourable recognition capabilities compared to classical baselines, achieving an average accuracy of over 90% on benchmarks with significantly fewer trainable parameters. Theoretical analysis confirms that QSVDD has a comparable expressivity to classical counterparts while requiring only a fraction of the parameters. Furthermore, we demonstrate the first implementation of a quantum machine learning method for anomaly detection on a superconducting quantum processor. Specifically, we achieve an accuracy of over 80% with only 16 parameters on the device, providing initial evidence of QSVDD's practical viability in the noisy intermediate-scale quantum era and highlighting its significant reduction in parameter requirements.
Pandemic Drugs at Pandemic Speed: Accelerating COVID-19 Drug Discovery with Hybrid Machine Learning- and Physics-based Simulations on High Performance Computers
Mar 04, 2021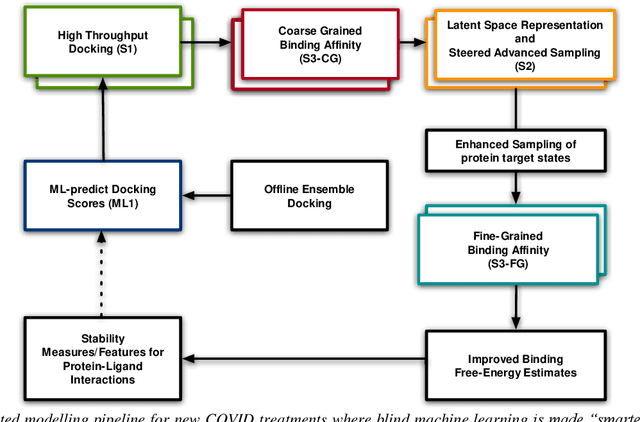
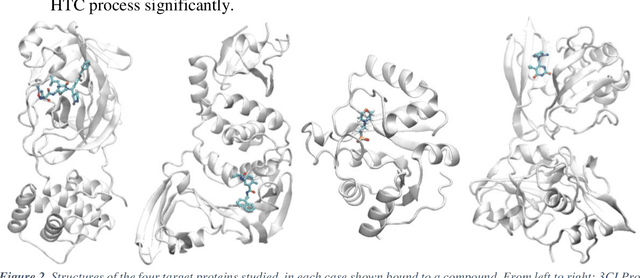
Abstract:The race to meet the challenges of the global pandemic has served as a reminder that the existing drug discovery process is expensive, inefficient and slow. There is a major bottleneck screening the vast number of potential small molecules to shortlist lead compounds for antiviral drug development. New opportunities to accelerate drug discovery lie at the interface between machine learning methods, in this case developed for linear accelerators, and physics-based methods. The two in silico methods, each have their own advantages and limitations which, interestingly, complement each other. Here, we present an innovative method that combines both approaches to accelerate drug discovery. The scale of the resulting workflow is such that it is dependent on high performance computing. We have demonstrated the applicability of this workflow on four COVID-19 target proteins and our ability to perform the required large-scale calculations to identify lead compounds on a variety of supercomputers.
HEMELB Acceleration and Visualization for Cerebral Aneurysms
Jun 27, 2019
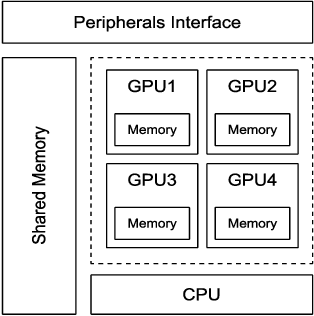
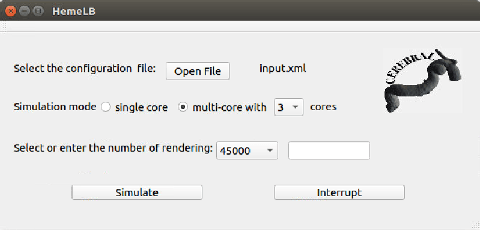
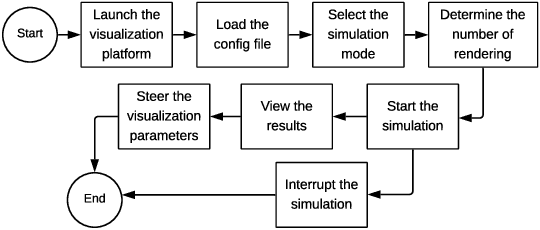
Abstract:A weakness in the wall of a cerebral artery causing a dilation or ballooning of the blood vessel is known as a cerebral aneurysm. Optimal treatment requires fast and accurate diagnosis of the aneurysm. HemeLB is a fluid dynamics solver for complex geometries developed to provide neurosurgeons with information related to the flow of blood in and around aneurysms. On a cost efficient platform, HemeLB could be employed in hospitals to provide surgeons with the simulation results in real-time. In this work, we developed an improved version of HemeLB for GPU implementation and result visualization. A visualization platform for smooth interaction with end users is also presented. Finally, a comprehensive evaluation of this implementation is reported. The results demonstrate that the proposed implementation achieves a maximum performance of 15,168,964 site updates per second, and is capable of speeding up HemeLB for deployment in hospitals and clinical investigations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge