Peter J. Schüffler
Deep Learning-Based Fixation Type Prediction for Quality Assurance in Digital Pathology
Feb 09, 2026Abstract:Accurate annotation of fixation type is a critical step in slide preparation for pathology laboratories. However, this manual process is prone to errors, impacting downstream analyses and diagnostic accuracy. Existing methods for verifying formalin-fixed, paraffin-embedded (FFPE), and frozen section (FS) fixation types typically require full-resolution whole-slide images (WSIs), limiting scalability for high-throughput quality control. We propose a deep-learning model to predict fixation types using low-resolution, pre-scan thumbnail images. The model was trained on WSIs from the TUM Institute of Pathology (n=1,200, Leica GT450DX) and evaluated on a class-balanced subset of The Cancer Genome Atlas dataset (TCGA, n=8,800, Leica AT2), as well as on class-balanced datasets from Augsburg (n=695 [392 FFPE, 303 FS], Philips UFS) and Regensburg (n=202, 3DHISTECH P1000). Our model achieves an AUROC of 0.88 on TCGA, outperforming comparable pre-scan methods by 4.8%. It also achieves AUROCs of 0.72 on Regensburg and Augsburg slides, underscoring challenges related to scanner-induced domain shifts. Furthermore, the model processes each slide in 21 ms, $400\times$ faster than existing high-magnification, full-resolution methods, enabling rapid, high-throughput processing. This approach provides an efficient solution for detecting labelling errors without relying on high-magnification scans, offering a valuable tool for quality control in high-throughput pathology workflows. Future work will improve and evaluate the model's generalisation to additional scanner types. Our findings suggest that this method can increase accuracy and efficiency in digital pathology workflows and may be extended to other low-resolution slide annotations.
From Linear Probing to Joint-Weighted Token Hierarchy: A Foundation Model Bridging Global and Cellular Representations in Biomarker Detection
Nov 07, 2025Abstract:AI-based biomarkers can infer molecular features directly from hematoxylin & eosin (H&E) slides, yet most pathology foundation models (PFMs) rely on global patch-level embeddings and overlook cell-level morphology. We present a PFM model, JWTH (Joint-Weighted Token Hierarchy), which integrates large-scale self-supervised pretraining with cell-centric post-tuning and attention pooling to fuse local and global tokens. Across four tasks involving four biomarkers and eight cohorts, JWTH achieves up to 8.3% higher balanced accuracy and 1.2% average improvement over prior PFMs, advancing interpretable and robust AI-based biomarker detection in digital pathology.
Deep Learning Methods for Lung Cancer Segmentation in Whole-slide Histopathology Images -- the ACDC@LungHP Challenge 2019
Aug 21, 2020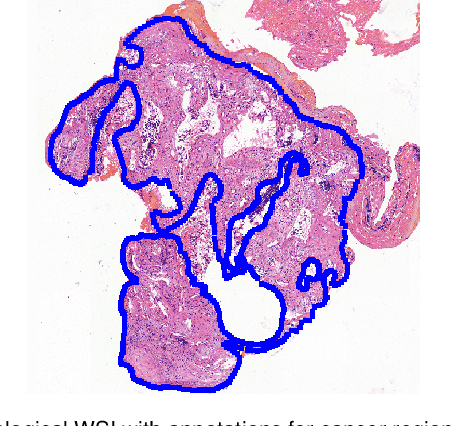
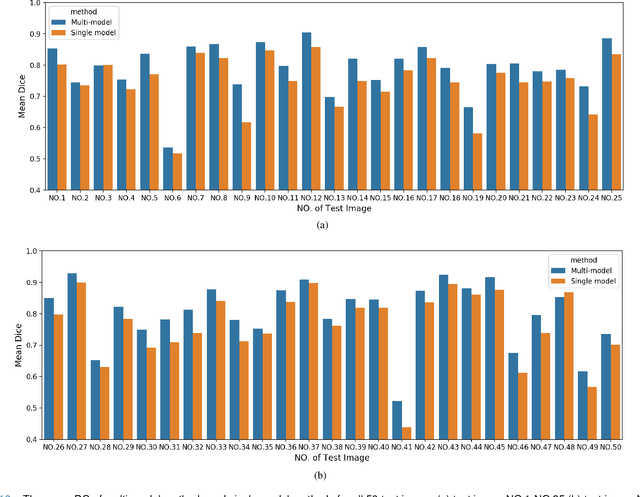
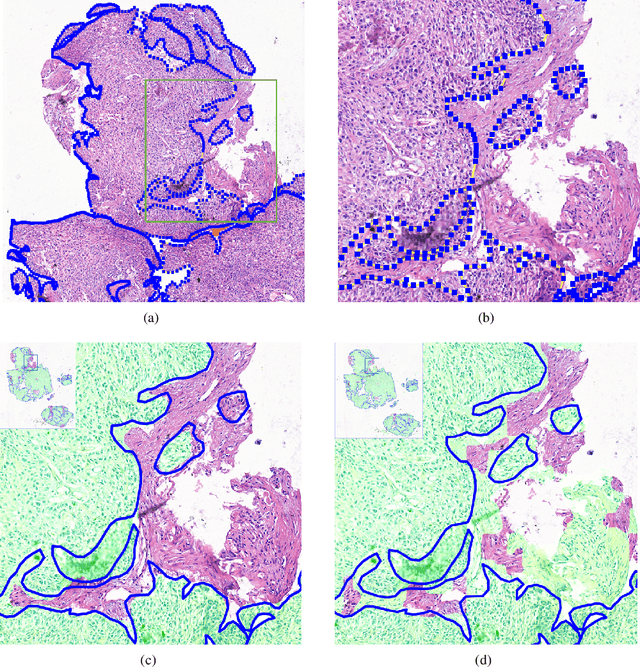
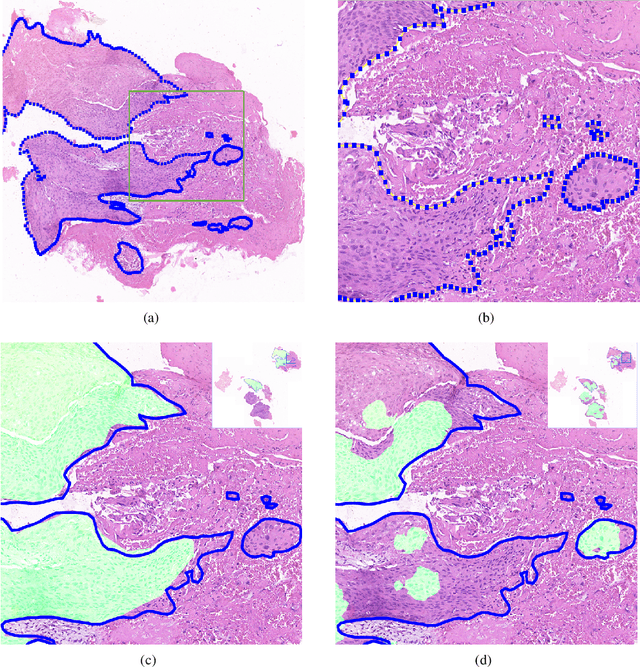
Abstract:Accurate segmentation of lung cancer in pathology slides is a critical step in improving patient care. We proposed the ACDC@LungHP (Automatic Cancer Detection and Classification in Whole-slide Lung Histopathology) challenge for evaluating different computer-aided diagnosis (CADs) methods on the automatic diagnosis of lung cancer. The ACDC@LungHP 2019 focused on segmentation (pixel-wise detection) of cancer tissue in whole slide imaging (WSI), using an annotated dataset of 150 training images and 50 test images from 200 patients. This paper reviews this challenge and summarizes the top 10 submitted methods for lung cancer segmentation. All methods were evaluated using the false positive rate, false negative rate, and DICE coefficient (DC). The DC ranged from 0.7354$\pm$0.1149 to 0.8372$\pm$0.0858. The DC of the best method was close to the inter-observer agreement (0.8398$\pm$0.0890). All methods were based on deep learning and categorized into two groups: multi-model method and single model method. In general, multi-model methods were significantly better ($\textit{p}$<$0.01$) than single model methods, with mean DC of 0.7966 and 0.7544, respectively. Deep learning based methods could potentially help pathologists find suspicious regions for further analysis of lung cancer in WSI.
Mitochondria-based Renal Cell Carcinoma Subtyping: Learning from Deep vs. Flat Feature Representations
Aug 02, 2016



Abstract:Accurate subtyping of renal cell carcinoma (RCC) is of crucial importance for understanding disease progression and for making informed treatment decisions. New discoveries of significant alterations to mitochondria between subtypes make immunohistochemical (IHC) staining based image classification an imperative. Until now, accurate quantification and subtyping was made impossible by huge IHC variations, the absence of cell membrane staining for cytoplasm segmentation as well as the complete lack of systems for robust and reproducible image based classification. In this paper we present a comprehensive classification framework to overcome these challenges for tissue microarrays (TMA) of RCCs. We compare and evaluate models based on domain specific hand-crafted "flat"-features versus "deep" feature representations from various layers of a pre-trained convolutional neural network (CNN). The best model reaches a cross-validation accuracy of 89%, which demonstrates for the first time, that robust mitochondria-based subtyping of renal cancer is feasible
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge