Pavan Ramdya
The embodied brain: Bridging the brain, body, and behavior with neuromechanical digital twins
Jan 12, 2026Abstract:Animal behavior reflects interactions between the nervous system, body, and environment. Therefore, biomechanics and environmental context must be considered to dissect algorithms for behavioral control. This is enabled by leveraging neuromechanical digital twins: computational models that embed artificial neural controllers within realistic body models in simulated environments. Here we review advances in the creation and use of neuromechanical digital twins while also highlighting emerging opportunities for the future. First, we illustrate how neuromechanical models allow researchers to infer hidden biophysical variables that may be difficult to measure experimentally. Additionally, by perturbing these models, one can generate new experimentally testable hypotheses. Next, we explore how neuromechanical twins have been used to foster a deeper exchange between neuroscience, robotics, and machine learning. Finally, we show how neuromechanical twins can advance healthcare. We envision that coupling studies on animals with active probing of their neuromechanical twins will greatly accelerate neuroscientific discovery.
Overcoming the Domain Gap in Neural Action Representations
Dec 23, 2021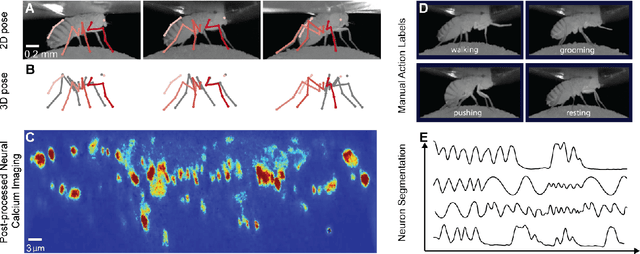
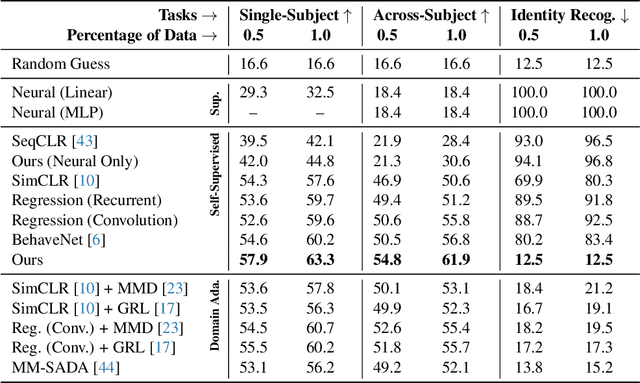
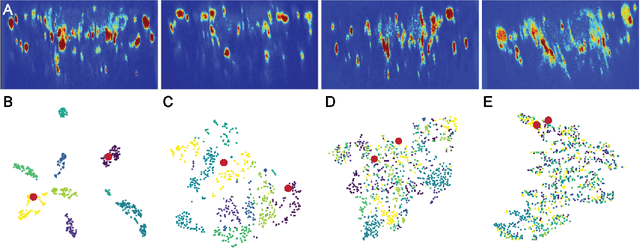
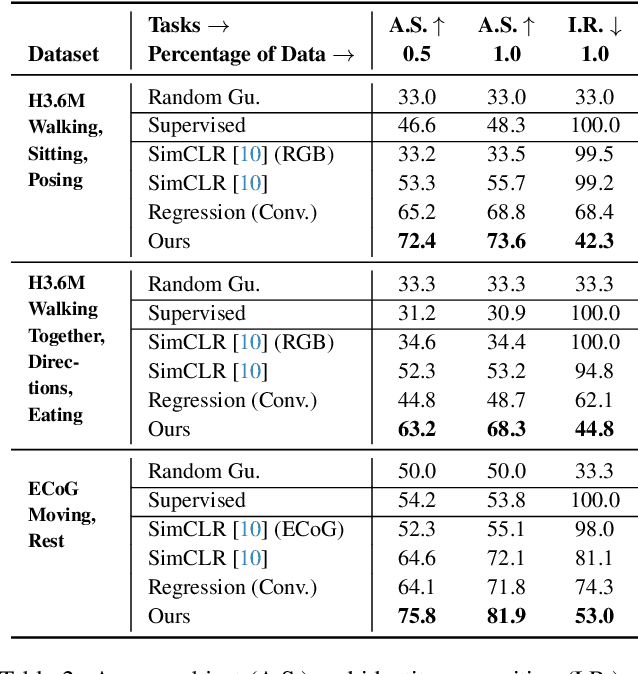
Abstract:Relating animal behaviors to brain activity is a fundamental goal in neuroscience, with practical applications in building robust brain-machine interfaces. However, the domain gap between individuals is a major issue that prevents the training of general models that work on unlabeled subjects. Since 3D pose data can now be reliably extracted from multi-view video sequences without manual intervention, we propose to use it to guide the encoding of neural action representations together with a set of neural and behavioral augmentations exploiting the properties of microscopy imaging. To reduce the domain gap, during training, we swap neural and behavioral data across animals that seem to be performing similar actions. To demonstrate this, we test our methods on three very different multimodal datasets; one that features flies and their neural activity, one that contains human neural Electrocorticography (ECoG) data, and lastly the RGB video data of human activities from different viewpoints.
Overcoming the Domain Gap in Contrastive Learning of Neural Action Representations
Nov 29, 2021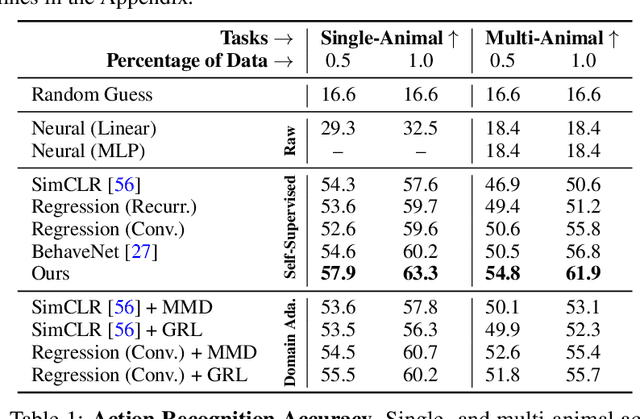
Abstract:A fundamental goal in neuroscience is to understand the relationship between neural activity and behavior. For example, the ability to extract behavioral intentions from neural data, or neural decoding, is critical for developing effective brain machine interfaces. Although simple linear models have been applied to this challenge, they cannot identify important non-linear relationships. Thus, a self-supervised means of identifying non-linear relationships between neural dynamics and behavior, in order to compute neural representations, remains an important open problem. To address this challenge, we generated a new multimodal dataset consisting of the spontaneous behaviors generated by fruit flies, Drosophila melanogaster -- a popular model organism in neuroscience research. The dataset includes 3D markerless motion capture data from six camera views of the animal generating spontaneous actions, as well as synchronously acquired two-photon microscope images capturing the activity of descending neuron populations that are thought to drive actions. Standard contrastive learning and unsupervised domain adaptation techniques struggle to learn neural action representations (embeddings computed from the neural data describing action labels) due to large inter-animal differences in both neural and behavioral modalities. To overcome this deficiency, we developed simple yet effective augmentations that close the inter-animal domain gap, allowing us to extract behaviorally relevant, yet domain agnostic, information from neural data. This multimodal dataset and our new set of augmentations promise to accelerate the application of self-supervised learning methods in neuroscience.
Deformation-aware Unpaired Image Translation for Pose Estimation on Laboratory Animals
Jan 23, 2020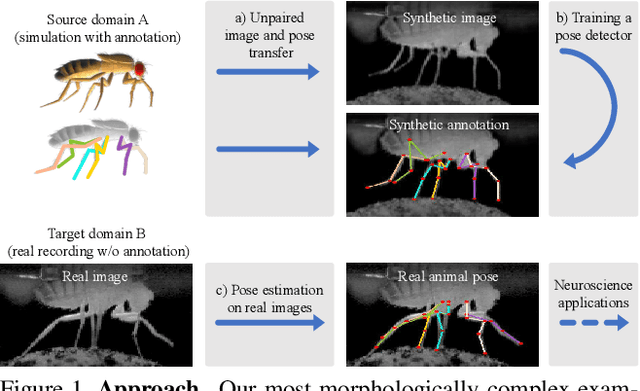
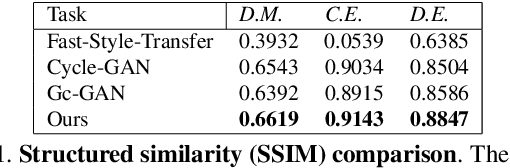
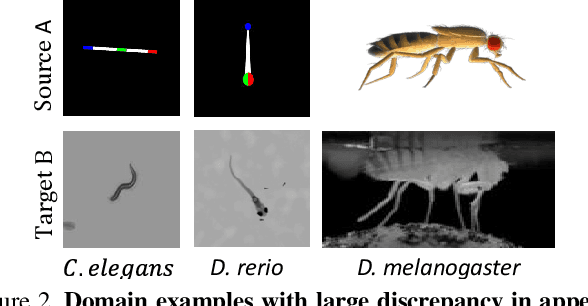
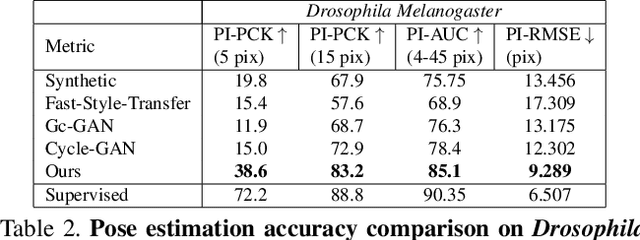
Abstract:Our goal is to capture the pose of neuroscience model organisms, without using any manual supervision, to be able to study how neural circuits orchestrate behaviour. Human pose estimation attains remarkable accuracy when trained on real or simulated datasets consisting of millions of frames. However, for many applications simulated models are unrealistic and real training datasets with comprehensive annotations do not exist. We address this problem with a new sim2real domain transfer method. Our key contribution is the explicit and independent modeling of appearance, shape and poses in an unpaired image translation framework. Our model lets us train a pose estimator on the target domain by transferring readily available body keypoint locations from the source domain to generated target images. We compare our approach with existing domain transfer methods and demonstrate improved pose estimation accuracy on Drosophila melanogaster (fruit fly), Caenorhabditis elegans (worm) and Danio rerio (zebrafish), without requiring any manual annotation on the target domain and despite using simplistic off-the-shelf animal characters for simulation, or simple geometric shapes as models. Our new datasets, code, and trained models will be published to support future neuroscientific studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge