Oya Aran
GIPSA-lab
Automated Measurement of Eczema Severity with Self-Supervised Learning
Apr 21, 2025Abstract:Automated diagnosis of eczema using images acquired from digital camera can enable individuals to self-monitor their recovery. The process entails first segmenting out the eczema region from the image and then measuring the severity of eczema in the segmented region. The state-of-the-art methods for automated eczema diagnosis rely on deep neural networks such as convolutional neural network (CNN) and have shown impressive performance in accurately measuring the severity of eczema. However, these methods require massive volume of annotated data to train which can be hard to obtain. In this paper, we propose a self-supervised learning framework for automated eczema diagnosis under limited training data regime. Our framework consists of two stages: i) Segmentation, where we use an in-context learning based algorithm called SegGPT for few-shot segmentation of eczema region from the image; ii) Feature extraction and classification, where we extract DINO features from the segmented regions and feed it to a multi-layered perceptron (MLP) for 4-class classification of eczema severity. When evaluated on a dataset of annotated "in-the-wild" eczema images, we show that our method outperforms (Weighted F1: 0.67 $\pm$ 0.01) the state-of-the-art deep learning methods such as finetuned Resnet-18 (Weighted F1: 0.44 $\pm$ 0.16) and Vision Transformer (Weighted F1: 0.40 $\pm$ 0.22). Our results show that self-supervised learning can be a viable solution for automated skin diagnosis where labeled data is scarce.
Ubiquitous Robot Control Through Multimodal Motion Capture Using Smartwatch and Smartphone Data
Jun 03, 2024



Abstract:We present an open-source library for seamless robot control through motion capture using smartphones and smartwatches. Our library features three modes: Watch Only Mode, enabling control with a single smartwatch; Upper Arm Mode, offering heightened accuracy by incorporating the smartphone attached to the upper arm; and Pocket Mode, determining body orientation via the smartphone placed in any pocket. These modes are applied in two real-robot tasks, showcasing placement accuracy within 2 cm compared to a gold-standard motion capture system. WearMoCap stands as a suitable alternative to conventional motion capture systems, particularly in environments where ubiquity is essential. The library is available at: www.github.com/wearable-motion-capture.
Visual In-Context Learning for Few-Shot Eczema Segmentation
Sep 28, 2023



Abstract:Automated diagnosis of eczema from digital camera images is crucial for developing applications that allow patients to self-monitor their recovery. An important component of this is the segmentation of eczema region from such images. Current methods for eczema segmentation rely on deep neural networks such as convolutional (CNN)-based U-Net or transformer-based Swin U-Net. While effective, these methods require high volume of annotated data, which can be difficult to obtain. Here, we investigate the capabilities of visual in-context learning that can perform few-shot eczema segmentation with just a handful of examples and without any need for retraining models. Specifically, we propose a strategy for applying in-context learning for eczema segmentation with a generalist vision model called SegGPT. When benchmarked on a dataset of annotated eczema images, we show that SegGPT with just 2 representative example images from the training dataset performs better (mIoU: 36.69) than a CNN U-Net trained on 428 images (mIoU: 32.60). We also discover that using more number of examples for SegGPT may in fact be harmful to its performance. Our result highlights the importance of visual in-context learning in developing faster and better solutions to skin imaging tasks. Our result also paves the way for developing inclusive solutions that can cater to minorities in the demographics who are typically heavily under-represented in the training data.
Inflammatory Bowel Disease Biomarkers of Human Gut Microbiota Selected via Ensemble Feature Selection Methods
Jan 08, 2020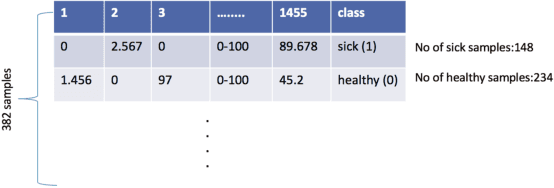
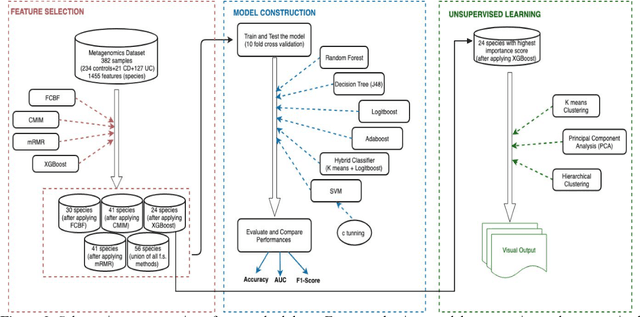
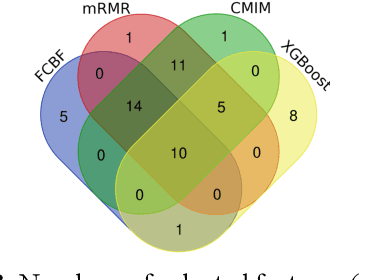
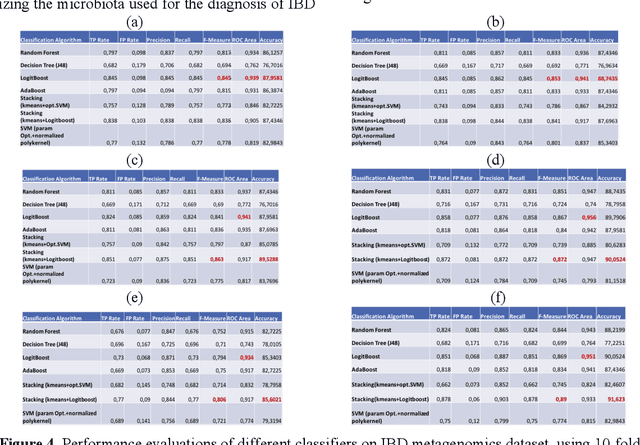
Abstract:The tremendous boost in the next generation sequencing and in the omics technologies makes it possible to characterize human gut microbiome (the collective genomes of the microbial community that reside in our gastrointestinal tract). While some of these microorganisms are considered as essential regulators of our immune system, some others can cause several diseases such as Inflammatory Bowel Diseases (IBD), diabetes, and cancer. IBD, is a gut related disorder where the deviations from the healthy gut microbiome are considered to be associated with IBD. Although existing studies attempt to unveal the composition of the gut microbiome in relation to IBD diseases, a comprehensive picture is far from being complete. Due to the complexity of metagenomic studies, the applications of the state of the art machine learning techniques became popular to address a wide range of questions in the field of metagenomic data analysis. In this regard, using IBD associated metagenomics dataset, this study utilizes both supervised and unsupervised machine learning algorithms, i) to generate a classification model that aids IBD diagnosis, ii) to discover IBD associated biomarkers, iii) to find subgroups of IBD patients using k means and hierarchical clustering. To deal with the high dimensionality of features, we applied robust feature selection algorithms such as Conditional Mutual Information Maximization (CMIM), Fast Correlation Based Filter (FCBF), min redundancy max relevance (mRMR) and Extreme Gradient Boosting (XGBoost). In our experiments with 10 fold cross validation, XGBoost had a considerable effect in terms of minimizing the microbiota used for the diagnosis of IBD and thus reducing the cost and time. We observed that compared to the single classifiers, ensemble methods such as kNN and logitboost resulted in better performance measures for the classification of IBD.
Sign Language Tutoring Tool
Feb 18, 2008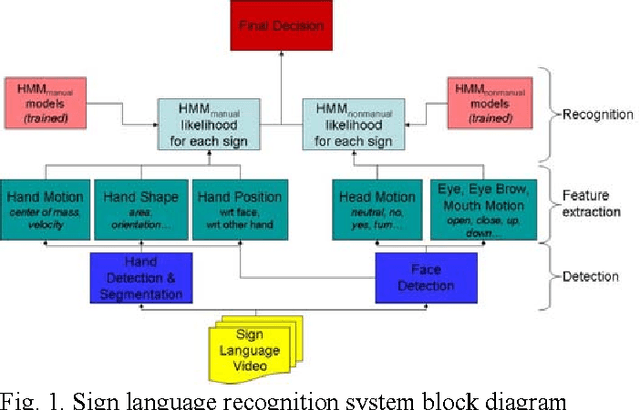
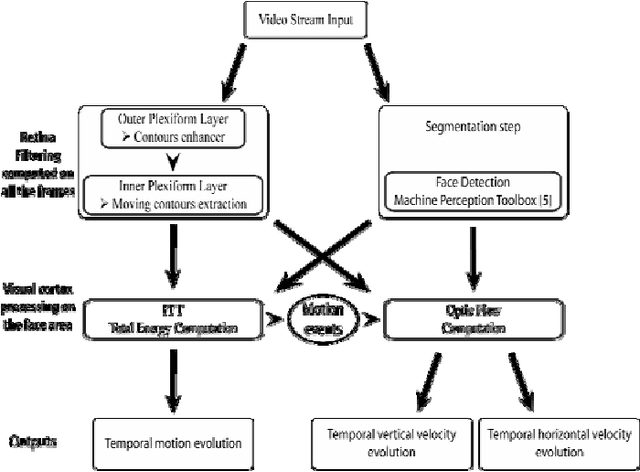
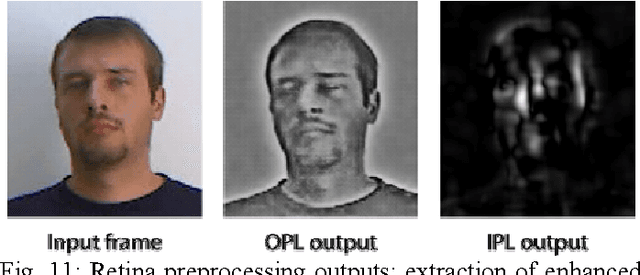
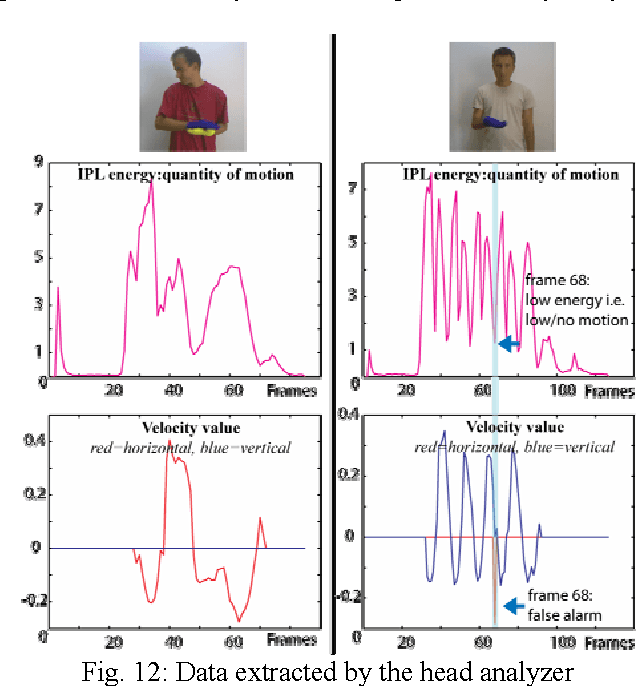
Abstract:In this project, we have developed a sign language tutor that lets users learn isolated signs by watching recorded videos and by trying the same signs. The system records the user's video and analyses it. If the sign is recognized, both verbal and animated feedback is given to the user. The system is able to recognize complex signs that involve both hand gestures and head movements and expressions. Our performance tests yield a 99% recognition rate on signs involving only manual gestures and 85% recognition rate on signs that involve both manual and non manual components, such as head movement and facial expressions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge