O. Ufuk Nalbantoglu
Residual Vision Transformer (ResViT) Based Self-Supervised Learning Model for Brain Tumor Classification
Nov 19, 2024
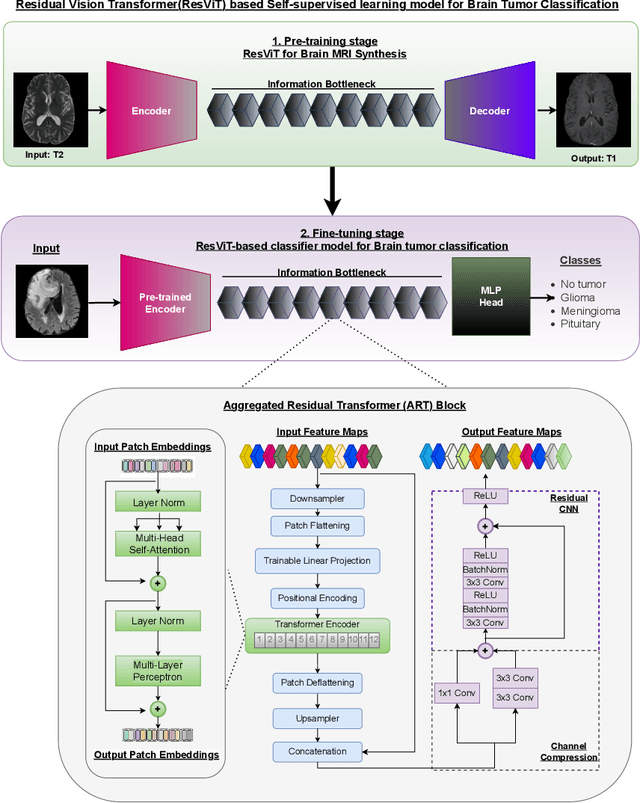
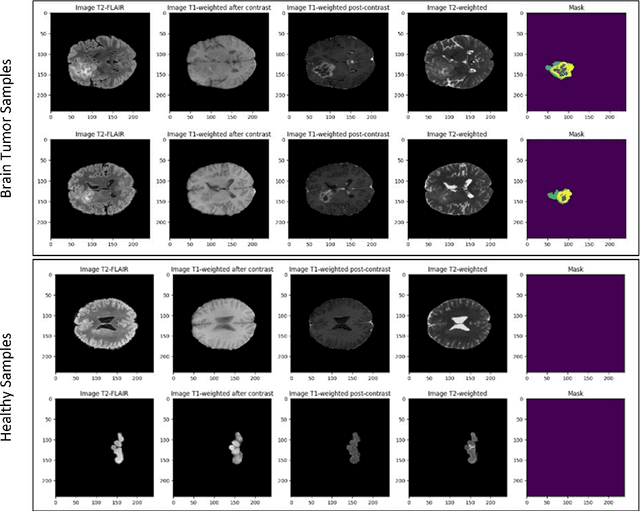
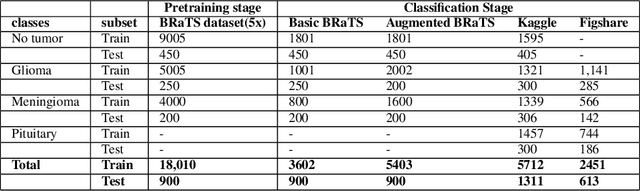
Abstract:Deep learning has proven very promising for interpreting MRI in brain tumor diagnosis. However, deep learning models suffer from a scarcity of brain MRI datasets for effective training. Self-supervised learning (SSL) models provide data-efficient and remarkable solutions to limited dataset problems. Therefore, this paper introduces a generative SSL model for brain tumor classification in two stages. The first stage is designed to pre-train a Residual Vision Transformer (ResViT) model for MRI synthesis as a pretext task. The second stage includes fine-tuning a ResViT-based classifier model as a downstream task. Accordingly, we aim to leverage local features via CNN and global features via ViT, employing a hybrid CNN-transformer architecture for ResViT in pretext and downstream tasks. Moreover, synthetic MRI images are utilized to balance the training set. The proposed model performs on public BraTs 2023, Figshare, and Kaggle datasets. Furthermore, we compare the proposed model with various deep learning models, including A-UNet, ResNet-9, pix2pix, pGAN for MRI synthesis, and ConvNeXtTiny, ResNet101, DenseNet12, Residual CNN, ViT for classification. According to the results, the proposed model pretraining on the MRI dataset is superior compared to the pretraining on the ImageNet dataset. Overall, the proposed model attains the highest accuracy, achieving 90.56% on the BraTs dataset with T1 sequence, 98.53% on the Figshare, and 98.47% on the Kaggle brain tumor datasets. As a result, the proposed model demonstrates a robust, effective, and successful approach to handling insufficient dataset challenges in MRI analysis by incorporating SSL, fine-tuning, data augmentation, and combining CNN and ViT.
Inflammatory Bowel Disease Biomarkers of Human Gut Microbiota Selected via Ensemble Feature Selection Methods
Jan 08, 2020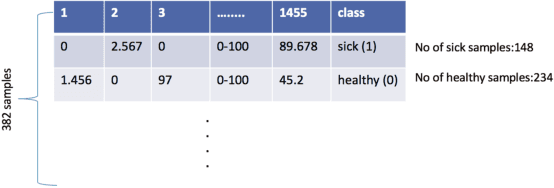
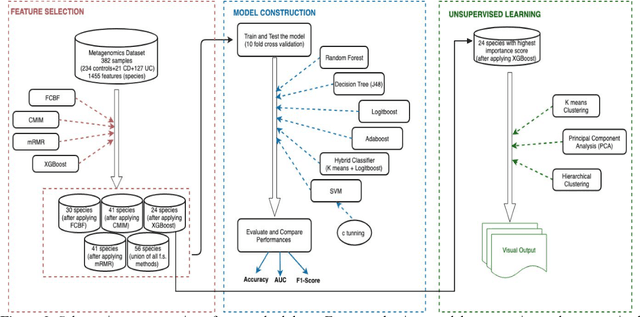
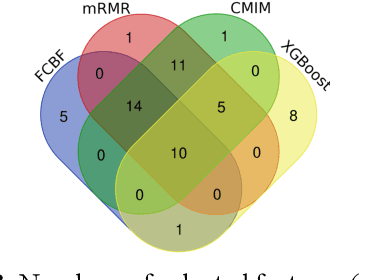
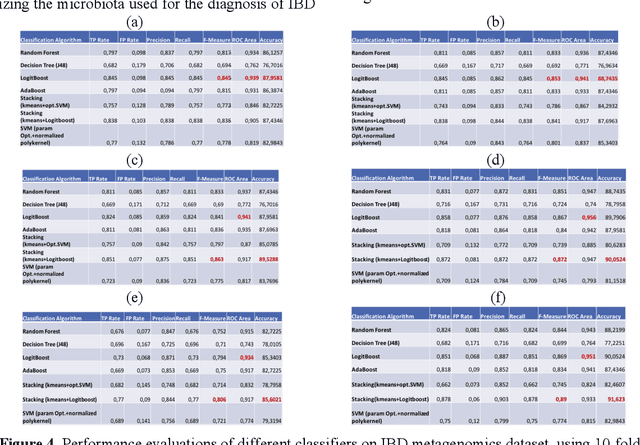
Abstract:The tremendous boost in the next generation sequencing and in the omics technologies makes it possible to characterize human gut microbiome (the collective genomes of the microbial community that reside in our gastrointestinal tract). While some of these microorganisms are considered as essential regulators of our immune system, some others can cause several diseases such as Inflammatory Bowel Diseases (IBD), diabetes, and cancer. IBD, is a gut related disorder where the deviations from the healthy gut microbiome are considered to be associated with IBD. Although existing studies attempt to unveal the composition of the gut microbiome in relation to IBD diseases, a comprehensive picture is far from being complete. Due to the complexity of metagenomic studies, the applications of the state of the art machine learning techniques became popular to address a wide range of questions in the field of metagenomic data analysis. In this regard, using IBD associated metagenomics dataset, this study utilizes both supervised and unsupervised machine learning algorithms, i) to generate a classification model that aids IBD diagnosis, ii) to discover IBD associated biomarkers, iii) to find subgroups of IBD patients using k means and hierarchical clustering. To deal with the high dimensionality of features, we applied robust feature selection algorithms such as Conditional Mutual Information Maximization (CMIM), Fast Correlation Based Filter (FCBF), min redundancy max relevance (mRMR) and Extreme Gradient Boosting (XGBoost). In our experiments with 10 fold cross validation, XGBoost had a considerable effect in terms of minimizing the microbiota used for the diagnosis of IBD and thus reducing the cost and time. We observed that compared to the single classifiers, ensemble methods such as kNN and logitboost resulted in better performance measures for the classification of IBD.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge